Validating Plant Gene Function: A Comprehensive Guide to VIGS and Knockout Studies
This article provides researchers and scientists with a detailed exploration of modern techniques for validating plant gene function, with a focus on Virus-Induced Gene Silencing (VIGS) and gene knockout studies.
Validating Plant Gene Function: A Comprehensive Guide to VIGS and Knockout Studies
Abstract
This article provides researchers and scientists with a detailed exploration of modern techniques for validating plant gene function, with a focus on Virus-Induced Gene Silencing (VIGS) and gene knockout studies. We cover foundational principles of post-transcriptional gene silencing, practical methodologies for implementing VIGS across diverse species including recalcitrant crops, and optimization strategies for enhancing silencing efficiency. The content includes direct comparisons between rapid VIGS screening and precise CRISPR/Cas9 genome editing, supported by case studies from recent research. Finally, we outline robust validation protocols and discuss the future integration of these tools for accelerating functional genomics and precision plant breeding.
The Science of Gene Silencing: From Natural Defense to Research Tool
Understanding Post-Transcriptional Gene Silencing (PTGS) as Plant Antiviral Defense
Post-Transcriptional Gene Silencing (PTGS) represents a fundamental RNA-based defense mechanism that plants employ to protect themselves against viral pathogens. This sequence-specific regulatory system recognizes and degrades aberrant or foreign RNA molecules, including viral genomes, providing an adaptive immune response that shares functional similarities with RNA interference (RNAi) pathways in other eukaryotes [1] [2]. The PTGS mechanism serves as a potent antiviral defense system because viral replication intermediates often generate double-stranded RNA (dsRNA) molecules, which the plant recognizes as "non-self" and targets for destruction [1]. This process effectively limits viral replication and spread within infected tissues, making PTGS a crucial component of plant innate immunity.
The significance of PTGS extends beyond its natural defensive role, as researchers have harnessed this mechanism to develop powerful functional genomics tools. Virus-Induced Gene Silencing (VIGS), which utilizes recombinant viral vectors to trigger targeted silencing of endogenous plant genes, has emerged as a particularly valuable application [3] [2]. The technology leverages the plant's own PTGS machinery to degrade both viral RNA and complementary host transcripts, enabling rapid functional characterization of genes involved in development, stress responses, and metabolic pathways [3] [4]. This review examines the molecular mechanisms of PTGS as an antiviral defense system, explores viral counter-defense strategies, and discusses experimental approaches for studying gene function through VIGS technology.
Molecular Mechanisms of Antiviral PTGS
The antiviral PTGS pathway operates through a well-orchestrated sequence of events that begins with detection of foreign RNA and culminates in sequence-specific degradation of target molecules. The process can be divided into distinct stages, each mediated by specific enzymatic complexes and signaling components.
Initiation: Detection and Dicing of Viral RNA
The PTGS pathway initiates when the plant detects double-stranded RNA (dsRNA) molecules, which are common replication intermediates for many viruses or can form through intramolecular base pairing within viral genomes [1] [5]. These dsRNA structures serve as potent signaling molecules recognized by host Dicer-like (DCL) enzymes, which belong to the RNase III family [1]. In Arabidopsis thaliana, four DCL enzymes have been identified, with DCL4 serving as the primary processor of virus-derived siRNAs, while DCL2 contributes to generating 22-nucleotide viral siRNAs and DCL3 produces 24-nucleotide variants involved in transcriptional silencing [1] [5]. The dicing activity of DCL enzymes cleaves long dsRNA molecules into small interfering RNA (siRNA) duplexes of 21-24 nucleotides in length, with the specific size distribution depending on the DCL enzyme involved [1].
Figure 1: Core Mechanism of Antiviral PTGS Pathway. The process begins with detection of viral dsRNA, progresses through siRNA-guided target cleavage, and amplifies through RDR6-dependent secondary siRNA production for systemic protection.
Effector Phase: RISC Assembly and Target Cleavage
Following dicing, the resulting virus-derived small interfering RNAs (vsiRNAs) are incorporated into the RNA-induced silencing complex (RISC), which serves as the catalytic heart of the silencing machinery [1] [5]. The slicing core of RISC contains Argonaute (AGO) proteins, which bind the vsiRNAs and use them as guides to identify complementary viral RNA sequences [1]. Arabidopsis possesses ten AGO proteins, with AGO1 playing a central role in antiviral defense, supported by AGO2 and AGO7 under specific conditions [1]. The programmed RISC complex scans viral transcripts and cleaves those complementary to the loaded vsiRNA guide strand, effectively suppressing viral replication and gene expression [1]. This sequence-specific degradation prevents translation of viral proteins, thereby limiting infection progression and symptom development.
Amplification and Systemic Signaling
Plants have evolved an amplification mechanism to enhance and sustain the PTGS response against viral pathogens. This amplification involves RNA-dependent RNA polymerases (RDRs), particularly RDR6, which use the cleavage products from primary RISC activity as templates to synthesize secondary dsRNA molecules [1] [5]. This process, stabilized by Suppressor of Gene Silencing 3 (SGS3), generates additional dsRNA substrates for DCL processing, resulting in production of secondary vsiRNAs that dramatically amplify the silencing signal [5]. Furthermore, the silencing signal spreads systemically throughout the plant, moving from cell to cell via plasmodesmata and over long distances through the vasculature, enabling establishment of antiviral immunity in distal tissues before viral invasion [5]. This systemic dimension of PTGS provides preemptive protection to non-infected tissues and represents a crucial adaptive feature of the plant immune system.
Viral Suppressors of RNA Silencing (VSRs)
In the ongoing evolutionary arms race between plants and viruses, viruses have developed sophisticated counter-defense strategies to overcome PTGS. Nearly all plant viruses encode viral suppressors of RNA silencing (VSRs)—multifunctional proteins that efficiently inhibit various steps of the antiviral silencing pathway [1] [5]. These suppressors employ diverse mechanisms to block PTGS, including dsRNA binding, siRNA sequestration, interference with AGO function, and inhibition of RDR activity [1]. The functional diversity of VSRs is remarkable, with even related viruses often employing structurally distinct proteins to achieve the same goal of silencing suppression [1].
Table 1: Characterized Viral Suppressors of RNA Silencing (VSRs)
| Viral Suppressor | Virus Origin | Mechanism of Action | Experimental Evidence |
|---|---|---|---|
| P1/HC-Pro | Tobacco etch virus | Suppresses PTGS at posttranscriptional level; nuclear transcription unaffected [6] | Stable expression in transgenic plants reversed silencing of GUS transgene [6] |
| 2b | Cucumber mosaic virus | Inhibits antiviral RNAi; interacts with AGO proteins [5] | Patch assays show restored reporter expression when co-expressed [1] |
| P19 | Tombusvirus | Binds and sequesters siRNAs to prevent RISC loading [1] | Crystal structures show siRNA duplex binding; prevents systemic silencing [1] |
| γb | Poa semilatent virus | Multifunctional; inhibits PTGS and regulates replication [1] | Genetic studies demonstrate silencing suppression activity [1] |
The classic "patch assay" has been instrumental in identifying and characterizing VSR activities [1]. This experimental approach involves co-expressing a candidate viral protein alongside a silencing inducer (e.g., dsRNA hairpin) and a reporter gene target. When a functional VSR is present, it prevents the silencing mechanism from degrading the reporter mRNA, resulting maintained reporter expression and minimal siRNA accumulation [1]. In contrast, absence of a functional suppressor leads to efficient reporter degradation and abundant siRNA production. This assay system has revealed that VSRs typically target core components of the PTGS machinery, with many exhibiting multiple mechanisms of inhibition to ensure robust suppression of host defenses [1].
Virus-Induced Gene Silencing (VIGS): Harnessing PTGS for Functional Genomics
VIGS represents a powerful application of PTGS that enables researchers to silence endogenous plant genes by engineering viral vectors to carry host-derived sequences. When these recombinant viruses infect plants, the PTGS machinery targets both viral RNA and the corresponding host mRNA, resulting in specific knockdown of the plant gene [3] [2] [4]. This technology has emerged as a versatile reverse genetics tool that bypasses the need for stable transformation, allowing rapid functional characterization of genes in a wide range of plant species [3] [2].
VIGS Vector Systems and Applications
Several RNA and DNA viruses have been engineered as VIGS vectors, each with distinct advantages and host range specificities. Tobacco Rattle Virus (TRV)-based vectors have gained particular popularity due to their broad host range, efficient systemic movement, and ability to target meristematic tissues [3] [7]. Other commonly used vectors include Potato Virus X (PVX), Tobacco Mosaic Virus (TMV), Cucumber Green Mottle Mosaic Virus (CGMMV), and DNA viruses like Geminiviruses [3] [2] [4]. The selection of an appropriate vector depends on the host plant species, target tissue, and duration of silencing required.
Table 2: Comparison of Major VIGS Vector Systems
| Vector System | Virus Type | Host Range | Key Features | Demonstrated Applications |
|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) | RNA virus | Broad (Solanaceae, Arabidopsis, etc.) | Efficient root & meristem invasion; mild symptoms [3] [7] | Silencing root development genes (IRT1, TTG1); nematode resistance (Mi) [7] |
| TRV-2b | Modified TRV | Extended range | Enhanced root tropism; improved meristem invasion [7] | Root architecture studies; nematode resistance pathways [7] |
| Cucumber Green Mottle Mosaic Virus (CGMMV) | RNA virus | Cucurbits (Luffa, cucumber, etc.) | Host-adapted for cucurbit species [4] | Tendril development (TEN); photobleaching phenotypes (PDS) [4] |
| Potato Virus X (PVX) | RNA virus | Solanaceous species | Strong systemic movement; robust silencing [2] | Disease resistance pathways; metabolic engineering [2] |
The practical implementation of VIGS involves cloning a fragment (typically 200-500 bp) of the target plant gene into a viral vector, introducing the construct into plants via Agrobacterium-mediated infiltration (agroinfiltration), mechanical inoculation, or other delivery methods, and monitoring for development of silencing phenotypes [3] [4]. Marker genes like Phytoene desaturase (PDS) and Cloroplastos Alterados 1 (CLA1) are commonly used to optimize VIGS protocols, as their silencing produces visible photobleaching phenotypes that confirm system functionality [8] [4]. For species with challenging leaf surfaces, such as Lycoris species with waxy coatings, modified infiltration methods like "leaf tip needle injection" have been developed to improve efficiency [8].
Figure 2: VIGS Experimental Workflow. The process involves constructing recombinant viral vectors, delivering them into plants, and analyzing resulting gene silencing phenotypes and molecular changes.
Experimental Validation and Research Applications
The functional validity of VIGS as a tool for gene characterization has been demonstrated across diverse plant species and biological processes. In pepper (Capsicum annuum L.), VIGS has enabled identification of genes governing fruit quality traits like color, biochemical composition, and pungency, as well as resistance to biotic and abiotic stresses [3]. In tomato, VIGS has been used to dissect disease resistance pathways and identify genes involved in defense signaling [2]. The technology has been particularly valuable for studying essential genes that would be lethal if constitutively disrupted, as VIGS produces transient knockdowns rather than permanent knockouts [2].
Recent methodological advances have further expanded VIGS applications. For root biology studies, a modified TRV vector retaining the 2b helper protein has shown dramatically improved efficiency in silencing genes in root tissues and meristems of Nicotiana benthamiana, Arabidopsis, and tomato [7]. This modified system enabled functional analysis of genes involved in root development (IRT1, TTG1, RHL1), lateral root-meristem function (RML1), and nematode resistance (Mi) [7]. In cucurbit species, CGMMV-based VIGS has been successfully established for ridge gourd (Luffa acutangula), silencing both marker genes (PDS) and developmental genes (TEN) involved in tendril formation [4]. These applications highlight how vector optimization and species-specific protocol development continue to broaden the utility of VIGS for plant functional genomics.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for PTGS and VIGS Studies
| Reagent / Material | Function / Application | Examples & Specifications |
|---|---|---|
| TRV-Based Vectors | Bipartite RNA virus system for VIGS | TRV1 (replicase/movement); TRV2 (capsid/target insert) [3] [7] |
| Marker Gene Constructs | VIGS efficiency validation | PDS (photobleaching); CLA1 (chloroplast development) [8] [4] |
| Agrobacterium Strains | Vector delivery via agroinfiltration | GV3101, others with appropriate antibiotic resistance [8] [4] |
| Viral Suppressor Clones | PTGS mechanism studies | P19, HC-Pro, 2b for suppression assays [1] [6] |
| siRNA Detection Reagents | Monitoring silencing efficiency | Northern blotting, small RNA sequencing protocols [1] [5] |
| AGO-Specific Antibodies | RISC complex immunoprecipitation | AGO1, AGO2 for studying slicing activity [1] [5] |
Post-Transcriptional Gene Silencing represents a sophisticated and highly adaptable antiviral defense system that plants have evolved to combat viral pathogens. The mechanistic understanding of this RNA-based immune response has not only revealed fundamental aspects of plant-virus interactions but has also enabled development of powerful research tools like VIGS that accelerate functional genomics. Despite viral counter-defense strategies through VSRs, the PTGS pathway remains a cornerstone of plant immunity, with ongoing research continuing to uncover novel regulatory components and signaling integrations. The experimental frameworks and reagent systems described herein provide researchers with robust methodologies for investigating gene function and dissecting molecular pathways in a wide range of plant species, thereby supporting advances in both basic plant science and applied agricultural biotechnology.
Virus-Induced Gene Silencing (VIGS) is a powerful reverse genetics technology that leverages the plant's innate antiviral RNA silencing machinery to suppress endogenous gene expression. As a sequence-specific post-transcriptional gene silencing (PTGS) mechanism, VIGS allows researchers to rapidly investigate gene function by introducing recombinant viral vectors carrying target plant gene fragments, leading to systemic silencing and observable phenotypic changes [3]. The molecular foundation of VIGS lies in the conserved RNA interference (RNAi) pathway, where double-stranded RNA (dsRNA) triggers are processed into small interfering RNAs (siRNAs) that guide the degradation of complementary mRNA sequences [9] [10]. This mechanism represents an adaptive defense system that plants have evolved to protect their genomes from invading nucleic acids, including viral pathogens [10] [11].
The significance of VIGS in modern plant research stems from its advantages over traditional stable transformation methods. VIGS offers a transient, rapid, and cost-effective approach for gene function characterization without the need for labor-intensive stable transformation, making it particularly valuable for plant species that are recalcitrant to genetic transformation [3] [12]. Since its initial demonstration in 1995 using a Tobacco mosaic virus vector carrying a phytoene desaturase (PDS) gene fragment, VIGS has been adapted for functional gene analysis in over 50 plant species, including major crops like tomato, barley, soybean, and cotton [3]. This technology has become an indispensable tool for high-throughput functional screening, especially in species like pepper (Capsicum annuum L.) where stable transformation remains challenging and genotype-dependent [3].
Molecular Machinery of VIGS
Core Components and Mechanisms
The molecular mechanism of VIGS operates through a sophisticated protein-RNA machinery that can be divided into distinct biochemical stages. The process begins when viral replicative double-stranded RNA (dsRNA) forms are recognized by the plant's silencing apparatus. These dsRNA intermediates, generated during viral replication, serve as the initial substrates for the RNAi pathway [9] [12]. The plant's Dicer-like (DCL) enzymes, which are RNase III family nucleases, then process these long dsRNA molecules into small interfering RNAs (siRNAs) of specific lengths [9] [10]. In Arabidopsis thaliana, four DCL enzymes (DCL1-4) generate different siRNA size classes: 21 nucleotides (nt) for DCL1 and DCL4, 22 nt for DCL2, and 24 nt for DCL3 [10].
These virus-derived siRNAs are incorporated into the RNA-induced silencing complex (RISC), a multiprotein effector complex that uses the siRNA as a guide to identify and cleave complementary viral RNA targets [9]. The core catalytic component of RISC is an Argonaute (AGO) protein, which possesses endonuclease activity ("slicer" activity) that executes the cleavage of target mRNAs [9] [11]. Arabidopsis possesses ten AGO proteins (AGO1-10) that show specialized functions in different silencing pathways [10]. The silencing effect becomes systemic through the movement of silencing signals between cells, allowing the entire plant to mount a defense against viral infection [3].
Table 1: Core Protein Components in Plant RNA Silencing Pathways
| Protein | Family | Function in VIGS | Specialized Roles |
|---|---|---|---|
| DCL1 | RNase III | Processes miRNA precursors | Generates 21-nt miRNAs from stem-loop structures |
| DCL2 | RNase III | Generates 22-nt siRNAs from viral dsRNA | Backup for DCL4; involved in secondary siRNA amplification |
| DCL3 | RNase III | Produces 24-nt heterochromatic siRNAs | Primarily involved in transcriptional silencing |
| DCL4 | RNase III | Primary processor of viral dsRNA | Generates 21-nt siRNAs for post-transcriptional silencing |
| AGO1 | Argonaute | Main effector for miRNA and siRNA function | Binds miRNAs and siRNAs to cleave complementary targets |
| AGO2 | Argonaute | Antiviral defense against certain viruses | Loads with viral siRNAs; enhanced expression upon infection |
| AGO7 | Argonaute | Binds specific miRNAs (e.g., miR390) | Involved in tasiRNA biogenesis |
| RDR6 | RNA-dependent RNA polymerase | Amplifies silencing signal | Converts cleaved RNAs into dsRNA for secondary siRNA production |
Visualizing the Core VIGS Pathway
The following diagram illustrates the key molecular steps in the VIGS mechanism, from viral infection to target gene silencing:
siRNA Biogenesis and Diversity
The siRNA population generated during VIGS exhibits considerable diversity in biogenesis pathways and functions. The major classes of small RNAs involved in silencing include:
Primary siRNAs: Derived directly from DCL processing of viral dsRNA replicative forms [9] [10]. These 21-24 nt molecules show asymmetric distribution along positive and negative viral RNA strands, with cleavage "hot spots" identified in tombusvirus infections [9].
Secondary siRNAs: Produced through an amplification mechanism involving RNA-dependent RNA polymerases (RDRs). When primary RISC complexes cleave target RNAs, the cleavage products can be converted to dsRNA by RDR6 (in Arabidopsis), which is then processed by DCL proteins into secondary siRNAs [10]. This process, known as "transitivity," enables amplification and systemic spread of the silencing signal [10].
microRNAs (miRNAs): Endogenous small RNAs processed from stem-loop precursor transcripts that can also be incorporated into RISC complexes. While not directly involved in VIGS, miRNAs share common machinery components and can influence silencing efficiency [9] [10].
The biogenesis of these different small RNA classes involves specialized protein complexes. For instance, the production of trans-acting siRNAs (tasiRNAs) requires specific RNA-binding proteins like SGS3, which interacts with RDR6 and may shuttle between nucleus and cytosol to facilitate RNA export and siRNA production [10].
Experimental Approaches for VIGS Mechanism Analysis
Key Methodologies and Protocols
Studying the molecular mechanism of VIGS requires specialized experimental approaches that can capture the dynamic protein-RNA interactions and enzymatic processes involved. The following core methodologies have been developed to dissect VIGS mechanisms:
Recombinant Viral Vector Construction is the foundational step in VIGS experiments. For tombusvirus-based systems like CymRSV, researchers clone 190-300 bp target gene fragments into viral genomes downstream of reporter genes like GFP [9]. The viral vector is then transformed into Agrobacterium tumefaciens strain GV3101 for plant delivery [12]. Optimal insert size and positioning within the viral genome significantly impact silencing efficiency, with studies using site-directed mutagenesis to create precise cloning sites [9].
Agrobacterium-Mediated Delivery (Agroinfiltration) involves growing recombinant Agrobacterium cultures to OD600 of 0.6-0.8 in YEP medium containing appropriate antibiotics, followed by resuspension in infiltration buffer (10 mM MgCl2, 10 mM MES, 200 μM AS) [12]. The bacterial suspension (OD600 0.8-1.0) is infiltrated into plant leaves using needleless syringes, typically targeting seedlings at the 4-6 leaf stage [9] [12]. Efficient delivery requires maintaining plants under high humidity with clear polyethylene covers for 24 hours post-infiltration [12].
Biochemical Analysis of RISC Complexes utilizes large-scale biochemical purification from plant tissues to isolate functional silencing complexes. The protocol involves homogenizing 100 mg of leaf tissue, followed by differential centrifugation to obtain cell extracts [9]. RISC complexes are purified through multiple chromatographic steps while monitoring for Ago-2 protein and RISC activity via Western blotting and nuclease assays [11]. Identification of associated proteins like VIG and dFXR (the Drosophila homolog of FMRP) is achieved through co-immunoprecipitation and RNAse treatment experiments [11].
High-Throughput Sequencing of Small RNAs provides comprehensive profiles of viral siRNAs. Researchers extract total RNA using Tri-reagent, then separate and detect small RNAs using 32P-labeled riboprobes or locked nucleic acid (LNA) oligonucleotide probes [9]. For precise mapping of cleavage sites, 3' rapid amplification of cDNA ends (3' RACE) sequencing is performed on 5 μg of total RNA ligated to 3'-end adapter oligonucleotides [9].
Table 2: Key Experimental Parameters for VIGS Analysis
| Parameter | Optimal Conditions | Impact on Results |
|---|---|---|
| Plant developmental stage | 4-6 leaf stage (Nicotiana benthamiana) | Younger plants show more efficient systemic silencing |
| Agroinfiltration OD600 | 0.8-1.0 | Higher OD can cause phytotoxicity; lower OD reduces efficiency |
| Post-infiltration temperature | 22-24°C constant | Elevated temperatures can enhance silencing spread but may increase non-specific effects |
| Time course analysis | 10-15 days post-inoculation | Early timepoints capture initiation; later timepoints show systemic effects |
| Tissue sampling | Separate local and systemic leaves | Distinguishes cell-autonomous from non-cell-autonomous silencing |
| siRNA detection method | High-resolution Northern blotting or small RNA-seq | Different sensitivity and quantification capabilities |
Visualizing the Experimental Workflow
The standard experimental pipeline for analyzing VIGS mechanisms involves both in vivo and in vitro approaches:
Key Research Reagents and Solutions
The following essential research materials are critical for successful investigation of VIGS mechanisms:
Table 3: Essential Research Reagents for VIGS Mechanism Studies
| Reagent/Solution | Composition/Specifications | Experimental Function |
|---|---|---|
| pTRV1 and pTRV2 Vectors | Bipartite Tobacco Rattle Virus system | Most versatile VIGS system for Solanaceae plants; TRV1 encodes replicase and movement proteins; TRV2 contains cloning site for target genes [3] |
| CymRSV-Cym19stop Mutant | Tombusvirus with defective p19 silencing suppressor | Unique system that allows strong VIGS activation in recovered leaves; enables analysis of RISC-mediated cleavage without suppressor interference [9] |
| CGMMV-based pV190 Vector | Cucumber green mottle mosaic virus vector | Specialized system for cucurbit species (cucumber, melon, Luffa); enables VIGS in previously challenging species [12] |
| Agrobacterium GV3101 | Strain with rifampicin and kanamycin resistance | Preferred strain for plant transformation; optimized for efficient T-DNA transfer in VIGS protocols [12] |
| Infiltration Buffer | 10 mM MgCl₂, 10 mM MES, 200 μM acetosyringone | Maintains Agrobacterium viability and promotes T-DNA transfer during leaf infiltration [12] |
| Tri-reagent Solution | Guanidine thiocyanate-phenol-chloroform mixture | Simultaneous extraction of RNA, DNA, and proteins from same sample; preserves small RNA species [9] |
| 32P-Labeled Riboprobes | In vitro transcribed RNAs with 32P-UTP | High-sensitivity detection of viral RNAs and siRNAs in Northern blot analyses [9] |
| LNA Oligonucleotide Probes | Locked Nucleic Acid-modified DNA oligos | Enhanced hybridization affinity for miRNA detection in Northern blots [9] |
| 3'-RACE Adapter Oligos | Pre-adenylated DNA/RNA chimeric oligonucleotides | Ligation to RNA 3' ends for amplification and sequencing of cleavage products [9] |
Comparative Analysis of VIGS Systems
Vector System Performance
Different viral vectors exhibit distinct performance characteristics in VIGS applications:
Table 4: Comparative Performance of Major VIGS Vector Systems
| Vector System | Host Range | Silencing Efficiency | Duration | Key Applications |
|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) | Broad (Solanaceae, Arabidopsis) | High in meristematic tissues | 3-5 weeks | Functional genomics in model plants; meristem gene silencing [3] |
| Tombusvirus (CymRSV) | Nicotiana benthamiana | Very high in recovered leaves | 2-4 weeks | Molecular mechanism studies; RISC activity analysis [9] |
| Cucumber Green Mottle Mosaic Virus (CGMMV) | Cucurbits (cucumber, Luffa, melon) | Moderate to high | 2-3 weeks | Gene function in cucurbit crops; fruit development studies [12] |
| Apple Latent Spherical Virus (ALSV) | Very broad (including legumes) | Variable between species | 4-8 weeks | Cross-species functional screening [12] |
| Bean Pod Mottle Virus (BPMV) | Soybean and other legumes | High in soybean | 3-6 weeks | Legume functional genomics [3] |
The TRV-based system remains the most widely adopted VIGS system due to its broad host range, efficient systemic movement, and ability to target meristematic tissues [3]. However, for specific plant families, specialized vectors like CGMMV for cucurbits have shown superior performance. Recent studies successfully applied CGMMV-VIGS to silence the tendril development gene TEN in Luffa acutangula, resulting in plants with shorter tendrils and higher nodal positions where tendrils appear [12]. Quantitative RT-PCR analysis confirmed significant reduction of TEN expression in silenced plants, demonstrating the efficacy of this system [12].
Molecular Efficacy Data
Experimental studies have generated quantitative data on the molecular efficacy of VIGS mechanisms:
Table 5: Quantitative Molecular Data on VIGS Efficacy
| Parameter | Experimental Measurement | Biological Significance |
|---|---|---|
| siRNA size distribution | 21-24 nucleotides (primary), 21-22 nt (secondary) | Reflects activities of different DCL enzymes; 21-nt siRNAs most abundant [9] [10] |
| Cleavage efficiency | Minus-sense RNA strands cleaved more efficiently than plus-sense | Asymmetric RISC activity may reflect accessibility differences in structured viral RNAs [9] |
| Silencing suppression | p19 protein binds 21-nt ds-siRNAs with high affinity | Viral counter-defense mechanism prevents siRNA incorporation into RISC [9] |
| RISC complex size | ~500 kD molecular mass | Indicates multiprotein composition including AGO, VIG, and dFXR proteins [11] |
| Temporal progression | Maximum siRNA accumulation at 10-15 dpi | Correlates with recovery phenotype in suppressor mutant viruses [9] |
| Spatial distribution | Higher silencing efficiency in vascular tissues | Suggests directional movement of silencing signal [9] |
Research using the CymRSV silencing suppressor mutant (Cym19stop) revealed that viral RNA targeting occurs primarily through RISC-mediated cleavage rather than translational inhibition [9]. This conclusion was supported by sensor construct experiments showing sequence-specific cleavage of viral target sequences but no evidence of translational repression [9]. Strikingly, these studies identified that RISC-mediated cleavages do not occur randomly on the viral genome but instead show hot spots with asymmetric distribution along positive and negative viral RNA strands [9].
Technical Considerations and Optimization
Critical Factors Affecting VIGS Efficiency
Several technical factors significantly influence the success and interpretation of VIGS experiments:
Plant Genotype and Developmental Stage profoundly impact VIGS efficiency. Studies in pepper (Capsicum annuum L.) reveal substantial genotype-dependent variation in silencing efficiency, with some cultivars showing robust systemic silencing while others exhibit only localized effects [3]. The optimal developmental stage for inoculation varies by species, but generally, younger plants (4-6 leaf stage for Nicotiana benthamiana) show more efficient systemic silencing [9] [12].
Environmental Conditions including temperature, humidity, and photoperiod must be carefully controlled. Maintaining constant temperature of 22-24°C following agroinfiltration significantly enhances silencing efficiency and reproducibility [9] [12]. High humidity immediately after infiltration is critical for Agrobacterium survival and T-DNA transfer, typically achieved by covering plants with clear polyethylene covers for 24-48 hours post-infiltration [12].
Agroinoculum Concentration requires precise optimization. While OD600 of 0.8-1.0 is commonly used, excessive Agrobacterium concentrations can cause phytotoxicity and non-specific effects, while insufficient concentrations reduce silencing efficiency [12]. The optimal concentration may need empirical determination for specific plant species and growth conditions.
Insert Design Parameters including length, sequence specificity, and positional effects within the viral vector significantly impact silencing efficiency. Fragments of 300-500 bp typically provide optimal specificity and efficacy, with GC content between 40-60% generally producing more consistent results [3] [12]. Bioinformatic analysis to avoid off-target silencing through sequence similarity searches is essential for accurate data interpretation.
Integration with Functional Genomics
VIGS serves as a powerful component in integrated functional genomics platforms, particularly when combined with other technologies. The marriage of VIGS with CRISPR/Cas9 systems enables rapid gene validation followed by precise genome editing [3] [13]. Similarly, integration with multi-omics approaches (transcriptomics, proteomics, metabolomics) provides comprehensive functional insights, as demonstrated in studies of manganese tolerance in mulberry where transcriptome analysis identified 811 differentially expressed genes under Mn stress, followed by VIGS validation of the MaCAX3 gene's role in Mn transport [14].
Recent advances in single-cell transcriptomic approaches further enhance the resolution of VIGS studies. Research in cotton used comparative single-cell transcriptomic mapping to identify a sea-island cotton-specific cell cluster, then employed VIGS to validate GbNF-YA7's role in pathogen resistance [15]. This integration of spatial resolution with functional validation represents the cutting edge of plant functional genomics.
The future of VIGS technology development points toward more sophisticated applications including virus-induced overexpression (VOX), virus-induced genome editing (VIGE), and host-induced gene silencing (HIGS) [12]. These advancements will further solidify VIGS as an indispensable tool for plant functional genomics and crop improvement programs.
For plant researchers, investigating gene function in recalcitrant species—those resistant to stable genetic transformation—presents a significant bottleneck. Traditional genetic engineering depends on efficient transformation and regeneration systems, which are absent in many legumes, woody perennials, and orphan crops [16] [17]. This limitation severely hinders functional genomics and the development of improved crop varieties. Fortunately, advanced transient techniques now enable rapid gene function analysis without the need for stable transformation. This guide compares three key approaches—Virus-Induced Gene Silencing (VIGS), Virus-Induced Genome Editing (VIGE), and Agrobacterium rhizogenes-mediated root transformation—providing researchers with actionable data and protocols to overcome these persistent challenges.
Technology Comparison & Performance Data
The following table objectively compares the core performance metrics of the three major technologies that bypass stable transformation.
Table 1: Performance Comparison of Technologies Bypassing Stable Transformation
| Technology | Primary Function | Key Advantage | Typical Efficiency | Time to Result | Key Limitation |
|---|---|---|---|---|---|
| VIGS [18] [19] [20] | Post-transcriptional gene silencing (knock-down) | Rapid phenotype generation; no plant transformation required | 60-83% silencing efficiency (varies by protocol) | 7-28 days post-infiltration | Transient; potential recovery from silencing |
| VIGE [21] [22] | Targeted genome editing (knock-out) | Production of transgene-free, heritable edits; bypasses tissue culture | Varies by virus and host; rapidly improving | Initial edits in first generation; homozygosity requires progeny | Limited cargo capacity; potential host immune reaction |
| A. rhizogenes Hairy Root [21] | Root-specific transformation | Enables functional study of genes in root biology; highly efficient in susceptible species | High transformation rates in compatible species (e.g., citrus, strawberry) | 2-4 weeks for root emergence | Limited to root tissues; not whole-plant editing |
Quantitative data demonstrates that VIGS is the fastest method for initial gene characterization, achieving high silencing efficiency within weeks. For instance, a 2023 study on Styrax japonicus established VIGS systems with silencing efficiencies of 83.33% (vacuum infiltration) and 74.19% (friction-osmosis) [18]. In Striga hermonthica, VIGS efficiency reached 60.2% via agro-infiltration [19]. VIGE, while potentially slower to achieve homozygous lines, offers the unique advantage of creating stable, transgene-free mutations that are heritable, which is crucial for crop improvement programs [22].
Detailed Experimental Protocols
VIGS Protocol for Recalcitrant Species
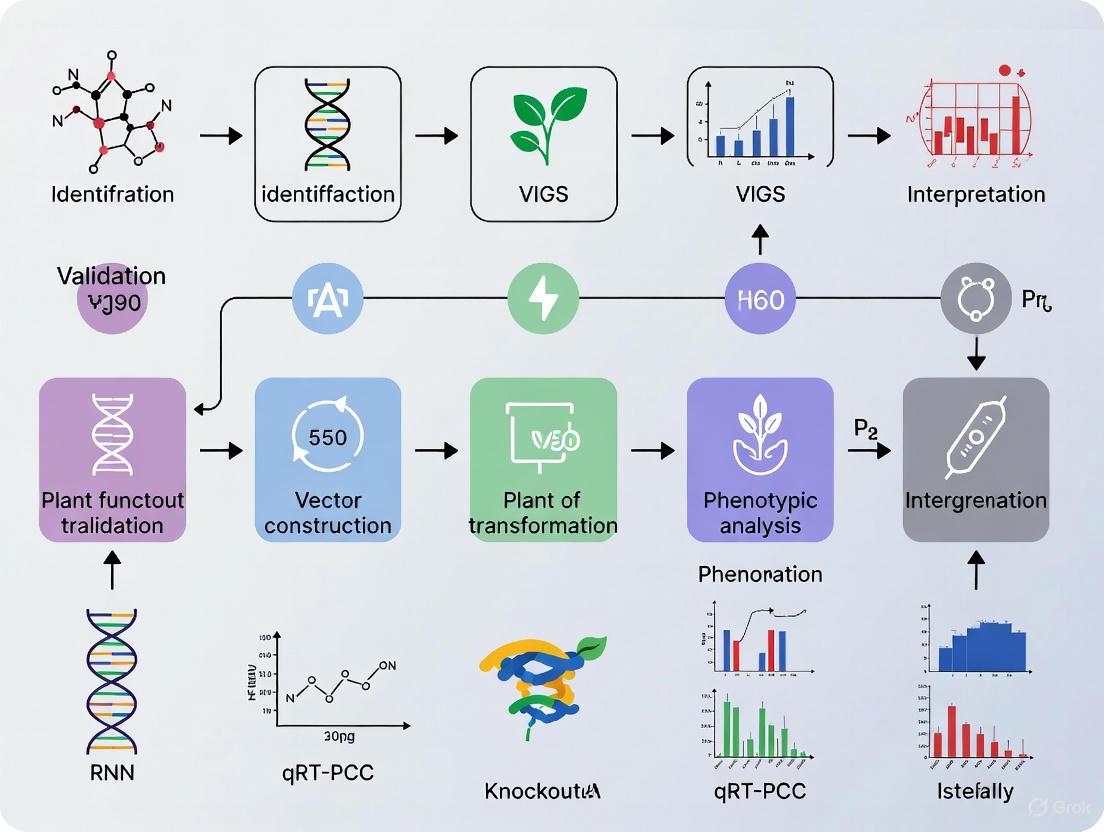
The Tobacco Rattle Virus (TRV)-based VIGS system is widely applicable. The following workflow and protocol detail its implementation.
Figure 1: VIGS Experimental Workflow
Step-by-Step Methodology:
- Vector Preparation: Clone a 300-500 bp fragment of the target gene (e.g., Phytoene desaturase (PDS) as a visual marker) into the multiple cloning site of the TRV2 vector. The construct is then introduced into Agrobacterium tumefaciens strains like GV3101 [19] [20].
- Agrobacterium Culture: Grow the Agrobacterium containing TRV1 and the recombinant TRV2 vectors in LB or TY media with appropriate antibiotics until the optical density at 600 nm (OD₆₀₀) reaches approximately 1.0 [19].
- Cell Preparation: Pellet the bacterial cells by centrifugation and resuspend them in an infiltration buffer containing 10 mM MgCl₂, 10 mM MES, and 200 μM acetosyringone to a final OD₆₀₀ of 0.5-1.0. The optimal density should be determined empirically for the target species [18] [21].
- Plant Infiltration:
- Agro-infiltration: Use a needleless syringe to infiltrate the bacterial suspension into the abaxial side of leaves for dicot species [19].
- Vacuum Infiltration: For whole-plant infiltration or difficult tissues, submerge seedlings or explants in the bacterial suspension and apply a vacuum for 2-5 minutes, followed by a slow release [18] [21].
- Agro-drench: Apply the bacterial suspension directly to the soil around the plant base, which is less efficient but minimally invasive [19].
- Plant Incubation and Analysis: Maintain infiltrated plants in high-humidity conditions for 1-2 days, then transfer to normal growth conditions. Silencing phenotypes (e.g., PDS photo-bleaching) typically appear 7 to 14 days post-infiltration. Validation requires molecular analysis like RT-qPCR using stable reference genes to quantify knockdown efficiency [18].
VIGE Protocol for Transgene-Free Editing
VIGE adapts VIGS principles for CRISPR/Cas9 genome editing.
Step-by-Step Methodology:
- System Configuration:
- Method A (Virus Delivering sgRNA): Use a viral vector (e.g., TRV, BSMV) to deliver the sgRNA sequence into transgenic plants that constitutively express the Cas9 protein [21].
- Method B (Virus Delivering Cas9 and sgRNA): Use a single or multiple viral vectors to deliver both Cas9 and sgRNA. This is limited by the cargo capacity of the virus and requires compact Cas9 variants or split systems [22].
- Viral Vector Inoculation: Inoculate plants with the engineered virus using methods similar to VIGS (agro-infiltration, rub-inoculation). The virus spreads systemically, delivering editing components to meristematic cells, which is crucial for heritable edits [22].
- Harvest and Genotyping: Allow the virus to spread for several weeks. Harvest seeds from inoculated plants (T₀). Screen the subsequent T₁ generation for edited alleles using PCR/RE assays or sequencing, as edits integrated into germline cells will be heritable [22].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for Bypassing Stable Transformation
| Reagent / Solution | Critical Function | Application Notes |
|---|---|---|
| TRV Vectors (pYL156, pYL279) [20] | RNA viral vector for inducing silencing; mild symptoms and robust systemic movement. | Preferred for Solanaceous species and Arabidopsis; efficient in meristems. |
| BSMV Vectors [20] | RNA viral vector for monocot species like barley and wheat. | Essential for silencing and editing studies in cereal crops. |
| Acetosyringone [18] [21] | Phenolic compound inducing Agrobacterium Vir genes; critical for T-DNA transfer. | Standard concentration of 200 μM in infiltration buffer is effective for many species. |
| Infiltration Buffer (MES/MgCl₂) [21] | Maintains pH and osmotic balance during Agrobacterium infiltration. | 10 mM MES (pH 5.2), 10 mM MgCl₂ provides optimal conditions. |
| Agrobacterium Strains (GV3101, K599) [21] [19] | Engineered disarmed strains for DNA delivery. GV3101 for VIGS/VIGE; K599 for hairy root induction. | K599 is a root-inducing (Ri) strain used for hairy root transformation. |
Bypassing stable transformation is no longer a barrier to functional genomics in recalcitrant plant species. VIGS stands out for its unparalleled speed in candidate gene validation, VIGE offers a direct path to heritable, transgene-free crop improvement, and A. rhizogenes-mediated transformation provides a unique window into root biology. The choice of technology depends entirely on the research objective: rapid knock-down, stable knock-out, or tissue-specific analysis. By adopting these transient and tissue culture-free methods, researchers can accelerate the characterization of gene function and the development of resilient crops, even in the most transformation-recalcitrant species.
Comparative Genomics and In Silico Prediction Models for Target Identification
In the modern genomics era, identifying causal genes and valid therapeutic targets requires sophisticated computational approaches that can process vast biological datasets. Comparative genomics and in silico prediction models have emerged as powerful methodologies for pinpointing critical genetic elements by analyzing evolutionary conservation, sequence patterns, and functional genomic data. These computational strategies are particularly valuable when integrated with experimental validation techniques like Virus-Induced Gene Silencing (VIGS) and CRISPR knockout screens, creating a robust framework for confirming gene function in plant systems and beyond [23] [3].
The fundamental premise of comparative genomics lies in identifying genetic elements conserved across species, which often indicate essential functional roles. Meanwhile, advanced machine learning models now leverage these comparative insights to predict the functional consequences of genetic variants with increasing accuracy. When deployed within a structured workflow, these computational tools enable researchers to efficiently prioritize candidate genes for expensive and time-consuming experimental validation, dramatically accelerating the pace of biological discovery and therapeutic development [23] [24].
Key In Silico Prediction Models and Performance
Model Architectures and Methodologies
In silico prediction models for genomic target identification primarily utilize two deep learning architectures: Convolutional Neural Networks (CNNs) and Transformer-based models. CNN-based approaches, including SEI and TREDNet, excel at identifying local sequence patterns such as transcription factor binding sites and chromatin features through their hierarchical feature extraction layers. These models process DNA sequences by applying filters that detect motif-like patterns, with successive layers integrating these patterns into higher-order regulatory signals [24].
Transformer-based architectures, such as DNABERT and Nucleotide Transformer, employ self-attention mechanisms to capture long-range dependencies in genomic sequences. These models are pre-trained on large-scale genomic datasets through self-supervised learning, allowing them to develop contextual understanding of DNA sequence functionality before being fine-tuned for specific prediction tasks like identifying enhancer variants or causal single-nucleotide polymorphisms (SNPs) [24].
Comparative Performance Analysis
Recent standardized benchmarking studies have provided critical insights into the relative strengths of different model architectures for specific genomic tasks. The table below summarizes the performance characteristics of leading models based on comprehensive evaluations across multiple datasets:
Table 1: Performance Comparison of Deep Learning Models for Genomic Predictions
| Model | Architecture | Best Application | Key Strengths | Performance Notes |
|---|---|---|---|---|
| SEI | CNN | Predicting regulatory impact of SNPs in enhancers | Captures local motif-level features effectively | Superior for enhancer variant effect prediction [24] |
| TREDNet | CNN | Predicting regulatory impact of SNPs in enhancers | Models local sequence motifs and regulatory elements | Excellent for estimating enhancer regulatory effects [24] |
| Borzoi | Hybrid CNN-Transformer | Causal variant prioritization within LD blocks | Integrates local and long-range sequence context | Best for identifying causal SNPs in linkage disequilibrium blocks [24] |
| DNABERT-2 | Transformer | Capturing long-range dependencies | Self-attention mechanisms for sequence context | Benefits significantly from fine-tuning [24] |
| Nucleotide Transformer | Transformer | Cell-type-specific regulatory effects | Pre-trained on large-scale genomic sequences | Performance improves with task-specific fine-tuning [24] |
For plant genomics applications, these models face additional challenges including large repetitive genomes, rapid functional turnover, and limited experimental data compared to mammalian systems. Nevertheless, they show strong potential for predicting variant effects in both coding and regulatory regions, extending traditional quantitative trait loci (QTL) mapping approaches by generalizing across genomic contexts rather than fitting separate models for each locus [23].
Experimental Validation Frameworks
Virus-Induced Gene Silencing (VIGS) in Plants
VIGS has emerged as a powerful reverse genetics tool for rapidly validating gene function in plants, leveraging the natural RNA silencing machinery of the host. The methodology involves engineering viral vectors to carry fragments of target plant genes, which when introduced into plants trigger sequence-specific degradation of complementary endogenous mRNAs through post-transcriptional gene silencing [3] [25].
The core VIGS workflow begins with vector selection and preparation, where researchers choose appropriate viral backbones (such as Tobacco Rattle Virus, Cucumber Green Mottle Mosaic Virus, or Turnip Crinkle Virus) and clone target gene fragments into the viral genome. The recombinant vectors are then introduced into Agrobacterium tumefaciens for plant transformation. For inoculation, Agrobacterium cultures carrying the VIGS constructs are infiltrated into plant tissues using needleless syringes, typically targeting leaves or cotyledons. After inoculation, plants are maintained under controlled environmental conditions to facilitate viral spread and silencing induction, with phenotypic effects typically observable within 2-4 weeks [12] [3] [25].
Table 2: Common VIGS Vectors and Their Applications in Plant Research
| Viral Vector | Virus Type | Host Range | Key Features | Example Applications |
|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) | RNA virus | Broad (Solanaceae, Arabidopsis) | Efficient systemic movement, mild symptoms | Functional genomics in pepper, tomato [3] |
| Cucumber Green Mottle Mosaic Virus (CGMMV) | RNA virus | Cucurbits (Luffa, cucumber) | Strong silencing in cucurbit species | Gene function studies in Luffa species [12] |
| Turnip Crinkle Virus (TCV) | RNA virus | Arabidopsis | Single RNA genome, high viral titer | Simultaneous silencing of multiple genes [25] |
| Apple Latent Spherical Virus (ALSV) | RNA virus | Broad (including cucurbits) | Mild or no symptoms | Silencing in multiple cucurbit species [12] |
Recent innovations in VIGS technology include the development of multiplex silencing vectors capable of simultaneously targeting multiple genes. For instance, researchers have engineered TCV-derived vectors that incorporate both a visual marker (e.g., phytoene desaturase, PDS) and additional target gene fragments, enabling preliminary assessment of silencing penetrance through visible photobleaching while studying genes of interest [25].
CRISPR/Cas9 Knockout Screens
CRISPR/Cas9-based functional screens represent a complementary approach to VIGS for target validation, particularly in systems where viral vectors are unsuitable. A notable advancement in this domain is the development of virus-free CRISPR screening methodologies that utilize plasmid-based sgRNA libraries coupled with whole-genome sequencing (WGS) for direct identification of causal mutations [26].
The experimental protocol involves several key steps. First, researchers design and synthesize a comprehensive sgRNA library targeting the protein-coding genes of interest, typically incorporating three sgRNAs per gene to ensure complete coverage. These plasmid libraries are co-transfected with a Cas9-expression plasmid into target cells, which are then subjected to selective pressures such as cytotoxic drugs or viral infection. Cells lacking key factors essential for survival under these conditions proliferate, while susceptible cells die. Genomic DNA from surviving cells is isolated and subjected to whole-genome sequencing to directly identify CRISPR/Cas9-induced causal mutations, bypassing statistical estimation approaches used in traditional lentiviral screens [26].
This approach has successfully identified both known and novel genes essential for viral infection in human cells, including the poliovirus receptor (PVR) and sialic acid biosynthesis genes (ST3GAL4) critical for enterovirus D68 infection. The methodology offers the distinct advantage of directly confirming causal mutations through WGS rather than relying on statistical enrichment of sgRNA sequences [26].
Integrated Workflows for Target Identification
Computational-Experimental Pipeline
Successful target identification requires careful integration of computational predictions with experimental validation. The workflow below illustrates the logical relationship between different stages of target identification:
Subtractive Genomics for Therapeutic Target Identification
In biomedical applications, subtractive genomics has emerged as a powerful computational strategy for identifying therapeutic targets in pathogenic organisms. This approach involves systematically comparing pathogen and host genomes to identify essential proteins in the pathogen that lack homologs in the host, thereby minimizing potential side effects from cross-reactivity [27].
The standard workflow begins with core proteome analysis to identify conserved proteins across multiple pathogen strains, followed by essentiality prediction using databases of essential genes. Non-essential genes and those with significant homology to host proteins are filtered out, leaving a subset of potential targets. Subsequent metabolic pathway analysis identifies proteins involved in pathogen-specific pathways, while subcellular localization predictions prioritize cytoplasmic targets for antibacterial development. Finally, druggability assessment evaluates the potential of shortlisted proteins to bind drug-like molecules with high affinity [27].
This methodology has successfully identified novel drug targets in multiple pathogens, including two proteins (B5ZC96 and B5ZAH8) in Ureaplasma urealyticum that show promise as therapeutic targets without significant homology to human proteins [27].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Genomic Target Identification and Validation
| Reagent/Tool | Category | Function | Example Applications |
|---|---|---|---|
| sgRNA Library | CRISPR Screening | Targets multiple genes for knockout | Genome-wide knockout screens [26] |
| Cas9 Expression Plasmid | CRISPR Screening | Provides DNA cleavage function | Co-transfection with sgRNA libraries [26] |
| TRV-Based VIGS Vectors | Plant Functional Genomics | Induces transient gene silencing | High-throughput gene validation in plants [3] |
| Agrobacterium tumefaciens GV3101 | Plant Transformation | Delivers genetic material into plants | VIGS vector delivery [12] |
| Whole Exome Sequencing Platforms | Genomics | Identifies coding region variants | Target discovery and validation [28] |
| Deep Learning Models (SEI, TREDNet) | Bioinformatics | Predicts regulatory variant effects | Prioritizing causal SNPs [24] |
The integration of comparative genomics, sophisticated in silico prediction models, and robust experimental validation frameworks has created a powerful paradigm for target identification across biological systems. As deep learning architectures continue to evolve and experimental methods become more precise, this integrated approach will undoubtedly accelerate the pace of discovery in both basic plant science and therapeutic development. The future of target identification lies in increasingly sophisticated computational models that can accurately predict gene function and variant impact across diverse biological contexts, coupled with high-throughput experimental methods that can rapidly validate these predictions at scale.
Virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics technology for rapidly characterizing gene functions in plants. This technology leverages the plant's innate antiviral RNA silencing defense mechanism to achieve sequence-specific downregulation of target genes without the need for stable genetic transformation. For cucurbit crops—economically important fruits and vegetables worldwide—functional genomic studies have been historically impeded by laborious and inefficient transformation protocols. The development of viral vectors capable of inducing gene silencing in cucurbits therefore represents a critical advancement for high-throughput gene function validation in these species. Among the various viral vectors available, Tobacco Rattle Virus (TRV), Cucumber Green Mottle Mosaic Virus (CGMMV), and Cucumber Fruit Mottle Mosaic Virus (CFMMV) have shown particular utility. This guide provides a comprehensive comparison of these three essential viral vectors, focusing on their host ranges, silencing efficiencies, and experimental applications to inform selection for functional genomics research.
Vector Origins and Genomic Features
Tobacco Rattle Virus (TRV)
TRV is a bipartite RNA virus belonging to the genus Tobravirus. Its genome consists of two RNA segments: RNA1 encoding replication and movement proteins, and RNA2 encoding the coat protein and other non-essential proteins that can be replaced with host gene fragments for VIGS. TRV has been widely adopted as a VIGS vector due to its broad host range, high silencing efficiency, and ability to induce long-lasting silencing effects in many plant species [29] [30]. The TRV-based VIGS system has been successfully established in tea plants (Camellia sinensis), where it silenced the CsPOR1 (protochlorophyllide oxidoreductase) gene, resulting in characteristic photobleaching phenotypes, and the CsTCS1 (caffeine synthase) gene, leading to a significant reduction in caffeine content [29].
Cucumber Green Mottle Mosaic Virus (CGMMV)
CGMMV is a positive-sense, single-stranded RNA virus belonging to the genus Tobamovirus. Its genome of approximately 6.4 kb contains four open reading frames encoding replication-associated proteins, a movement protein (MP), and a coat protein (CP) [31] [32]. CGMMV-based VIGS vectors have been developed by inserting multiple cloning sites or duplicating the CP subgenomic promoter to accommodate foreign gene fragments [31]. CGMMV naturally infects cucurbit plants and has been engineered as an effective VIGS vector for several cucurbit species, demonstrating mild viral symptoms and persistent silencing effects that can last for over two months [31] [30].
Cucumber Fruit Mottle Mosaic Virus (CFMMV)
CFMMV is also a member of the genus Tobamovirus with genomic organization similar to CGMMV. Recent vector development efforts have optimized CFMMV for VIGS applications by incorporating the Araujia mosaic virus (ArjMV) MP gene, which significantly enhanced its silencing efficiency and stability in cucurbit hosts [30]. The improved CFMMV vector achieves higher silencing efficiency and longer duration of gene silencing effects compared to earlier versions, making it particularly valuable for functional studies in cucurbits [30].
Comparative Host Range and Silencing Efficiency
Table 1: Host Range Comparison of TRV, CGMMV, and CFMMV VIGS Vectors
| Plant Species | TRV | CGMMV | CFMMV | Key Experimental Findings |
|---|---|---|---|---|
| Nicotiana benthamiana | Effective [29] | Effective [31] | Information Missing | CGMMV vector with duplicated CP subgenomic promoter (pV190) caused milder symptoms than wild-type virus [31] |
| Cucumber (Cucumis sativus) | Effective [30] | Effective [31] [4] | Effective [30] | CGMMV vector induced photobleaching by silencing PDS; TRV required special agroinfiltration solution [31] [30] |
| Watermelon (Citrullus lanatus) | Effective [30] | Effective [31] [4] | Effective [30] | CGMMV-based silencing persisted for over two months; CFMMV silenced genes related to male sterility [31] [30] |
| Melon (Cucumis melo) | Effective [30] | Effective [31] | Information Missing | TRSV and ALSV vectors have also been successfully used [30] |
| Bottle Gourd (Lagenaria siceraria) | Information Missing | Effective [31] [4] | Information Missing | CGMMV vector effectively silenced PDS gene [31] |
| Ridge Gourd (Luffa acutangula) | Information Missing | Effective [4] | Information Missing | CGMMV-VIGS system successfully silenced PDS and TEN genes, affecting tendril development [4] |
| Tea (Camellia sinensis) | Effective [29] | Information Missing | Information Missing | TRV-mediated silencing of CsPOR1 and CsTCS1 achieved with vacuum infiltration [29] |
Table 2: Performance Characteristics of VIGS Vectors in Cucurbit Species
| Vector | Silencing Duration | Key Advantages | Reported Limitations |
|---|---|---|---|
| TRV | Long-lasting [29] | Wide host range beyond cucurbits; Established protocols | May require optimization of infection methods for different cucurbits [30] |
| CGMMV | Over 2 months [31] | Natural cucurbit pathogen; High efficiency in multiple species; Mild symptoms with optimized vectors | Silencing efficiency varies with insert size and orientation [31] |
| CFMMV | Long-lasting [30] | High efficiency with optimized vector; Effective for floral trait studies | Limited application reports compared to other vectors [30] |
| TrMMV | Persistent [33] | Broad efficacy across cucurbits; Particularly high efficiency in C. melo; Useful for floral traits | Newer vector with less established protocols [33] |
Experimental Protocols and Methodologies
TRV-Mediated VIGS Protocol
The TRV-VIGS system typically employs a two-component vector system (pTRV1 and pTRV2). For tea plants, researchers have successfully implemented the following protocol [29]:
- Vector Construction: Clone target gene fragments (e.g., CsPOR1 or CsTCS1) into the pTRV2 vector.
- Agrobacterium Preparation: Transform recombinant pTRV2 and pTRV1 into Agrobacterium tumefaciens strains such as GV3101.
- Plant Inoculation: Mix bacterial suspensions containing pTRV1 and recombinant pTRV2 in a 1:1 ratio, then infiltrate into plants using vacuum infiltration or syringe infiltration.
- Phenotype Observation: Silencing phenotypes typically appear within 2-4 weeks post-inoculation.
In tea plants, this approach achieved approximately 75% silencing efficiency for CsPOR1, resulting in obvious photobleaching symptoms, and significantly reduced CsTCS1 expression, leading to a 6.26-fold decrease in caffeine content [29].
CGMMV-Based VIGS Protocol
The CGMMV-VIGS system has been optimized for cucurbit species including cucumber, watermelon, and ridge gourd [31] [4]:
- Vector Design: Develop CGMMV derivatives (e.g., pV190) with duplicated CP subgenomic promoters and BamHI cloning sites.
- Insert Cloning: Clone target gene fragments (e.g., PDS or TEN) in sense orientation into the vector.
- Agroinfiltration: Introduce recombinant vectors into Agrobacterium strain GV3101 and infiltrate into cotyledons or true leaves of seedlings.
- Efficiency Optimization: Use fragments of 150-300 bp for optimal silencing efficiency, with hairpin structures potentially enhancing silencing.
This system has successfully silenced the TEN gene in ridge gourd, resulting in altered tendril development with higher nodal positions of tendril appearance and shorter tendril length [4].
Impact of Insert Size on Silencing Efficiency
Research with the Trichosanthes mottle mosaic virus (TrMMV) VIGS system, a related tobamovirus, demonstrated that insert sizes between 90-400 bp can induce effective silencing, with 150 bp fragments showing particularly high efficiency in Cucumis sativus [33]. Similar size dependencies have been observed for CGMMV-based vectors, where fragments of 150-300 bp achieved optimal silencing [31].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for VIGS Experiments in Cucurbits
| Reagent/Resource | Function/Purpose | Examples/Specifications |
|---|---|---|
| VIGS Vectors | Delivery of target gene fragments into host plants | pTRV1/pTRV2 (TRV), pV190 (CGMMV), pCF93 (CFMMV) [29] [31] [30] |
| Agrobacterium Strains | Delivery of viral vectors into plant cells | GV3101, EHA105 [33] [4] |
| Marker Genes | Visual assessment of silencing efficiency | PDS (photobleaching), POR1 (chlorophyll synthesis), Su (chlorophyll biosynthesis) [33] [29] [31] |
| Infiltration Buffers | Facilitating Agrobacterium entry into plant tissues | 10 mM MgCl₂, 10 mM MES, 200 μM AS [4] |
| Detection Primers/Probes | Confirming viral infection and silencing efficiency | Coat protein, movement protein, or replicase-specific primers [34] |
TRV, CGMMV, and CFMMV each offer distinct advantages as VIGS vectors for plant functional genomics research. TRV provides the broadest host range, extending beyond cucurbits to species like tea plants, making it suitable for comparative studies across diverse plant families. CGMMV demonstrates superior performance in natural cucurbit hosts, with persistent silencing effects and mild symptom development in optimized vectors. CFMMV represents a specialized tool for cucurbits, particularly valuable for studying reproductive development and traits when using recently enhanced versions. Vector selection should be guided by target host species, experimental timeframe, and specific research objectives. For cucurbit-specific studies, CGMMV and CFMMV generally offer more reliable and efficient silencing, while TRV remains the vector of choice for broader host ranges or when working with non-cucurbit species. Continuing vector optimization, particularly in delivery methods and insert stability, will further enhance the utility of these tools for high-throughput functional genomics in recalcitrant plant species.
Practical Implementation: From Vector Design to Cross-Species Application
The validation of plant gene function relies heavily on reverse-genetics approaches, with Virus-Induced Gene Silencing (VIGS) emerging as a particularly powerful tool for transient gene knockdown in species recalcitrant to stable transformation. VIGS operates by harnessing the plant's endogenous post-transcriptional gene silencing (PTGS) machinery, using recombinant viral vectors to systemically suppress target gene expression. The efficacy of this technology is not merely a function of viral vector selection but is profoundly influenced by the strategic design and orientation of the inserted gene fragment. Within the broader context of plant functional genomics, where techniques range from RNA interference (RNAi) to CRISPR-Cas9-mediated knockout, VIGS offers a unique balance of speed and versatility. This guide objectively compares the performance of different viral vector systems and insert design strategies, providing supporting experimental data to inform researchers' selection and construction protocols for optimal gene functional validation.
Viral Vector Systems for VIGS: A Comparative Analysis
The choice of viral vector is a primary determinant of VIGS success, influencing host range, tissue tropism, and silencing efficiency. Different viral backbones offer distinct advantages and limitations.
Table 1: Comparison of Key Viral Vectors Used in VIGS
| Vector System | Genome Type | Key Features/Advantages | Limitations | Demonstrated Silencing Efficiency |
|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) [3] | RNA virus | Broad host range (especially Solanaceae); efficient systemic movement including meristems; mild symptoms. | Bipartite genome requires two vectors (TRV1, TRV2). | ~16.4% in Atriplex canescens (germinated seeds/vacuum infiltration) [35]; 40-80% transcript reduction in N. benthamiana roots with TRV-2b vector [7]. |
| Potato Virus X (PVX) [36] | RNA virus | Well-characterized genome; suitable for deconstruction. | Can cause severe viral symptoms; limited insert capacity (~2 kb). | GFP yield of 0.13 mg/g FW in N. benthamiana; 3-4 fold increase to 0.50 mg/g FW with integrated VSRs [36]. |
| Bean Yellow Dwarf Virus (BeYDV) [37] | DNA virus (Geminivirus) | High-level, transient gene expression; replication in plant nuclei. | Protein detection possible within 3-7 days post-infiltration [37]. | |
| Broad Bean Wilt Virus 2 (BBWV2) [3] | RNA virus | Effective in a range of crops, including pepper. | Widely used for functional genomics in Capsicum annuum [3]. |
The TRV-based system is often the vector of choice for Solanaceae plants like pepper and tomato due to its high efficiency and minimal pathology [3]. A key advancement in TRV vector design involves the retention of the RNA2-encoded 2b protein. Contrary to earlier constructs that deleted this gene, TRV vectors retaining the 2b protein demonstrate significantly enhanced invasion of root and meristematic tissues in Nicotiana benthamiana, Arabidopsis thaliana, and tomato, leading to a more robust and pervasive systemic VIGS response in these critical tissues [7]. This makes the TRV-2b vector indispensable for functional studies of genes involved in root development and soil-borne pathogen interactions.
The Role of Viral Suppressors of RNA Silencing (VSRs)
A significant limitation of viral vectors is their recognition and suppression by the host's RNA silencing machinery. Engineering vectors to co-express heterologous Viral Suppressors of RNA Silencing (VSRs) can dramatically enhance recombinant protein expression and, by extension, silencing potential.
Table 2: Enhancement of VIGS Efficiency via Heterologous VSRs
| VSR | Origin | Mechanism of Action | Experimental Vector | Performance Enhancement |
|---|---|---|---|---|
| P19 [36] | Tomato bushy stunt virus (TBSV) | Sequesters siRNAs to prevent RISC incorporation. | PVX-derived (pP2) | Markedly enhanced GFP fluorescence and accumulation in N. benthamiana [36]. |
| P38 [36] | Turnip crinkle virus (TCV) | Binds directly to AGO1 protein. | PVX-derived (pP2) | Strong enhancement of GFP expression, second only to NSs [36]. |
| NSs [36] | Tomato zonate spot virus (TZSV) | Targets SGS3 for degradation. | PVX-derived (pP2) | Highest GFP accumulation: 0.50 mg/g FW, a 3.8-fold increase over parental PVX vector [36]. |
A critical finding in vector construction is that the transcriptional orientation of the inserted VSR cassette relative to the target gene is a major factor in its efficacy. Initial constructs with VSR and target gene in the same orientation showed reduced expression, likely due to transcriptional interference. Simply reversing the orientation of the VSR cassette in the PVX-based vector (creating pP3-based constructs) alleviated this interference, leading to a significant boost in the expression of both the VSR and the target gene (e.g., GFP or vaccine antigens VP1 and S2) [36]. For vaccine antigens, this strategy resulted in yield increases of over 100-fold compared to the parental PVX vector [36].
Insert Design: Principles and Protocols
The design of the insert fragment cloned into the viral vector is equally critical for successful silencing.
- Fragment Selection and Size: The insert should be a highly specific, 300-400 base pair fragment derived from the coding sequence of the target gene [3] [35]. Online tools like the SGN-VIGS tool (https://vigs.solgenomics.net/) can be used to predict optimal nucleotide target regions and verify sequence specificity to avoid off-target silencing [35]. Fragments corresponding to the 5' end, central region, and 3' end of the open reading frame are typically tested, with the central fragment often yielding the highest efficiency [35].
- Avoiding Secondary Structure: The insert sequence should be analyzed to avoid regions with high potential for stable secondary structure, which can impede viral replication or the generation of siRNAs.
Experimental Protocol: Cloning and Vector Construction
- Gene Fragment Amplification: Amplify the selected target gene fragment (e.g., AcPDS, AcTIP2;1) from cDNA using primers containing appropriate restriction enzyme sites (e.g., EcoRI and BamHI) [35].
- Restriction Digestion and Ligation: Digest both the amplified PCR product and the VIGS vector (e.g., pTRV2) with the chosen restriction enzymes. Purify the digested fragments and ligate them together using T4 DNA ligase [35].
- Transformation and Verification: Transform the ligation product into competent E. coli cells. Select positive colonies, isolate plasmid DNA, and verify the correct insertion of the gene fragment via colony PCR and sequencing [35].
- Agrobacterium Transformation: Introduce the verified recombinant vector plasmid (e.g., TRV2:Target) and the complementary helper vector (e.g., TRV1) into Agrobacterium tumefaciens strain GV3101 using freeze-thaw transformation [35].
Diagram 1: VIGS Vector Construction Workflow. This diagram outlines the key steps in cloning a target gene fragment into a viral vector for VIGS.
Inoculation Methodologies and Optimization
The method of delivering the viral vector into the plant is a major experimental variable.
Table 3: Comparison of VIGS Inoculation Methods
| Method | Protocol Description | Key Parameters | Optimal Efficiency & Applications |
|---|---|---|---|
| Vacuum Infiltration [35] | Submerging germinated seeds/seedlings in Agrobacterium suspension and applying a vacuum. | 0.5 kPa for 5-10 min; OD600 = 0.8 [35]. | Highest efficiency: 16.4% in A. canescens; ideal for high-throughput silencing of germinated seeds [35]. |
| Syringe Infiltration [37] | Pressing Agrobacterium suspension into the abaxial side of leaves using a needleless syringe. | OD600 = 0.5-1.0; incubation in infiltration buffer (10 mM MES, 200 µM AS, 10 mM MgCl2) [3]. | Standard for leaf assays in model plants like N. benthamiana; suitable for protein expression within days [37]. |
| Soaking [35] | Immersing plant materials in Agrobacterium suspension with gentle shaking. | 40 min immersion; 50 rpm shaking [35]. | Lower efficiency compared to vacuum infiltration; a simpler, less equipment-intensive alternative [35]. |
The plant's developmental stage is also critical. Inoculation of germinated Atriplex canescens seeds with radicles of 1-3 cm resulted in successful systemic silencing, whereas inoculation of intact seeds with seed coats was ineffective [35]. Furthermore, environmental conditions post-inoculation, including temperature, humidity, and photoperiod, must be carefully controlled to optimize both plant health and viral spread, thereby maximizing silencing efficiency [3].
VIGS in the Context of Alternative Gene Silencing Technologies
While VIGS is a powerful knockdown tool, its position within the functional genomics toolkit is defined by comparison with other technologies.
- VIGS vs. RNAi: Both VIGS and transgenic RNAi cause gene knockdown at the mRNA level. However, VIGS is transient and does not require stable transformation, making it faster and applicable to non-transformable species [3]. A significant drawback of RNAi is its propensity for high off-target effects due to the silencing of mRNAs with limited complementarity [38].
- VIGS vs. CRISPR-Cas9: CRISPR-Cas9 creates permanent knockouts at the DNA level, allowing for complete and heritable gene disruption [38]. This is a key advantage for studying essential genes where partial knockdown may not reveal a phenotype. However, lethal knockouts can prevent functional analysis, whereas the transient, partial knockdown of VIGS can still provide insights [38]. CRISPR workflows are also generally longer and more dependent on efficient transformation protocols. Newer CRISPR applications like CRISPR interference (CRISPRi) can create reversible knockouts, blurring the line between the two approaches [38].
Diagram 2: Gene Silencing Technology Comparison. This chart compares the mechanism, effect, and key advantage of VIGS, RNAi, and CRISPR-Cas9.
Case Study: Validating a Root Development Gene
To illustrate the integrated application of these principles, consider a study aiming to validate the function of a putative aquaporin gene (AcPIP2;5) in the root system of Atriplex canescens [35].
- Vector Selection: A TRV-based vector is chosen for its documented efficiency in roots, especially the TRV-2b variant.
- Insert Design: A 428-bp specific fragment from the AcPIP2;5 ORF is selected using the SGN-VIGS tool and cloned into the pTRV2 vector to create TRV2:AcPIP2;5.
- Inoculation: Germinated A. canescens seeds are inoculated with the Agrobacterium suspension (OD600 = 0.8) carrying TRV1 and TRV2:AcPIP2;5 using vacuum infiltration (0.5 kPa, 10 min).
- Validation: Systemic silencing phenotypes appear in new leaves and roots around 15 days post-inoculation. qRT-PCR analysis confirms a 69.5% reduction in AcPIP2;5 transcript abundance, and the observed phenotype (e.g., altered root hydraulics) phenocopies the expected function, validating the gene's role [35].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagent Solutions for VIGS Experiments
| Reagent / Material | Function in VIGS Protocol | Example & Specification |
|---|---|---|
| VIGS Vectors | Engineered viral genomes to deliver and amplify the target gene insert. | pTRV1 and pTRV2 plasmids for TRV-based system [3] [35]. |
| Agrobacterium Strain | Bacterial vehicle for delivering the VIGS vector into plant cells. | A. tumefaciens GV3101 [35]. |
| Infiltration Buffer | Solution to prepare Agrobacterium for plant infiltration. | 10 mM MES, 200 µM Acetosyringone, 10 mM MgCl₂, 0.03% Silwet-77 [35]. |
| Marker Gene | Visual reporter for optimizing silencing efficiency and tracking viral spread. | Phytoene desaturase (PDS); silencing causes photobleaching [3] [35]. |
| Viral Suppressor of RNAi (VSR) | Enhances silencing efficiency by suppressing host defense. | NSs protein from Tomato zonate spot virus for maximum protein yield boost [36]. |
The selection and construction of vectors for VIGS are far from trivial steps, with decisions regarding viral backbone, insert design, orientation, and delivery method directly dictating the success of functional gene validation studies. As the data demonstrates, optimized TRV vectors, particularly those incorporating the 2b protein or heterologous VSRs like NSs in a reverse orientation, can push silencing efficiency to new heights, especially in challenging tissues like roots. When objectively compared to stable RNAi or CRISPR-based knockout, VIGS maintains its niche as the fastest and most accessible technique for transient gene knockdown in non-model plants. The continued refinement of vector design and inoculation protocols ensures that VIGS will remain an indispensable component of the plant functional genomics toolkit, crucial for accelerating gene discovery and crop breeding programs.
In the field of plant functional genomics, Agroinfiltration serves as a cornerstone technique for the transient introduction of genetic material into plant tissues, enabling rapid validation of gene function through approaches such as virus-induced gene silencing (VIGS) and knockout studies [3] [39]. This technique leverages the natural DNA transfer capability of Agrobacterium tumefaciens to deliver gene constructs directly into plant cells, bypassing the need for stable transformation [40]. Among the various delivery methods available, vacuum infiltration and seed soaking have emerged as two prominent, efficient protocols, each with distinct advantages and optimal applications for different plant species and experimental requirements. This guide provides an objective comparison of these methodologies, supported by experimental data, to assist researchers in selecting the most appropriate protocol for their functional genomics research.
Seed Soaking Method
The seed soaking method involves immersing germinated or non-germinated seeds in an Agrobacterium suspension for a predetermined duration to facilitate gene transfer. This approach is particularly valued for its technical simplicity and ability to generate whole-plant transformation systems.
- Typical Protocol: In a study on Paeonia ostii, researchers optimized a seed soaking protocol using in vitro embryo-derived seedlings. The optimal conditions identified were an Agrobacterium concentration (OD600) of 1.0, an immersion time of 2 hours, and the inclusion of 200 µM acetosyringone in the infiltration solution [41].
- Plant Material Preparation: The process often begins with sterile embryo excision. For tree peony, this involved inoculating embryos onto specific germination media (MS or WPM) and culturing them under controlled conditions (16h light/8h dark at 24±2°C) to produce consistent seedling materials [41].
Vacuum Infiltration Method
Vacuum infiltration employs negative pressure to remove air from plant tissues, allowing the Agrobacterium suspension to penetrate intercellular spaces more effectively than passive methods.
- Standard Procedure: For sunflower seedlings, an optimized ultrasonic-vacuum protocol involved first subjecting 3-day-old seedlings cultured in Petri dishes to ultrasonication at 40 kHz for 1 minute, followed by vacuum infiltration at 0.05 kPa for 5-10 minutes [40].
- Infiltration Solution Composition: Research in wheat and maize demonstrated the importance of infiltration solution composition. A novel solution containing acetosyringone, cysteine, and Tween 20 enabled whole-plant level VIGS when combined with vacuum infiltration of germinated seeds [42].
The workflow below illustrates the key steps involved in each method:
Comparative Performance Analysis
Efficiency and Transformation Rates
Both methods can achieve high transformation efficiency when optimized for specific plant species, though their performance varies across biological systems.
Table 1: Efficiency Comparison Between Vacuum Infiltration and Seed Soaking
| Plant Species | Method | Optimal Parameters | Reported Efficiency | Key Experimental Findings | Citation |
|---|---|---|---|---|---|
| Paeonia ostii (Tree Peony) | Seed Soaking | OD600=1.0, 200 µM AS, 2h infection | High transformation efficiency achieved at 35 days post-germination | Maximum transformation efficiency required 6 negative-pressure treatments during soaking | [41] |
| Sunflower | Vacuum Infiltration | 0.05 kPa, 5-10 min, 0.02% Silwet L-77 | >90% transformation efficiency | Gene expression sustained for at least 6 days post-infiltration | [40] |
| Sunflower | Seed Soaking (Infiltration) | OD600=0.8, 0.02% Silwet L-77, 2h immersion | >90% transformation efficiency | Prolonged immersion (>4h) caused tissue damage and root necrosis | [40] |
| Wheat & Maize | Vacuum Infiltration | Acetosyringone, cysteine, Tween 20 solution | Whole-plant level gene silencing | Enabled VIGS in monocots; silenced PDS and MLO genes effectively | [42] |
Applications in Functional Genomics
Both methods have been successfully applied to key functional genomics techniques:
VIGS Applications: The seed soaking method has been effectively used for TRV-mediated VIGS in tree peony to characterize gene functions through transient silencing and overexpression [41]. Similarly, vacuum infiltration has enabled whole-plant VIGS in wheat and maize, allowing functional analysis of genes involved in pathogen resistance and early plant development [42].
Protein Localization and Promoter Studies: Seed soaking protocols have been adapted for subcellular protein localization studies and investigating transcription factor-mediated regulation of target gene promoters using GFP and GUS reporter systems [41].
Abiotic Stress Research: Both methods facilitate rapid validation of genes involved in stress response pathways. For example, vacuum infiltration in sunflower enabled functional analysis of the HaNAC76 gene in response to salt and drought stress [40].
Detailed Experimental Protocols
Seed Soaking Protocol for Tree Peony
Adapted from the TTAES (Transient Transformation system mediated by Agrobacterium using in vitro embryo-derived seedlings) method [41]
Materials Preparation:
- Prepare embryo germination media (MS medium supplemented with 30 g/L sucrose, 0.5 mg/L 6-BA, and 1.0 mg/L GA3, pH 5.6)
- Surface-sterilize seeds through sequential washing with detergent, 75% ethanol (30s), and 2% sodium hypochlorite (2×6 min)
- Excise embryos aseptically and inoculate onto germination media
- Culture under 16h light/8h dark photoperiod at 24±2°C
Agroinfiltration Procedure:
- Grow Agrobacterium harboring the gene construct to OD600 = 1.0 in appropriate medium
- Add 200 µM acetosyringone to the bacterial suspension
- Immerse germinated seedlings (35 days after germination) in the bacterial suspension
- Apply negative pressure treatments (6 times) during the 2-hour infection period
- Transfer seedlings to fresh media and co-cultivate for 2-3 days
- Assess transformation efficiency using GUS staining or GFP visualization
Vacuum Infiltration Protocol for Sunflower
Adapted from established methods for efficient transient transformation [40]
Materials Preparation:
- Grow sunflower seedlings for 3 days in Petri dishes or hydroponically
- Prepare Agrobacterium strain GV3101 carrying the pBI121 vector with GUS reporter
- Adjust bacterial concentration to OD600 = 0.8 in infiltration solution
- Add 0.02% Silwet L-77 as surfactant to the bacterial suspension
Infiltration Procedure:
- Submerge seedlings completely in the Agrobacterium suspension
- Place the container in a vacuum desiccator
- Apply vacuum (0.05 kPa) for 5-10 minutes until air bubbles cease to emerge
- Gradually release the vacuum to allow thorough infiltration
- Remove seedlings from the suspension and blot excess liquid
- Co-cultivate infiltrated seedlings in the dark at room temperature for 3 days
- Analyze gene expression using histochemical GUS assays or molecular analysis
Research Reagent Solutions
Table 2: Essential Reagents for Agroinfiltration Protocols
| Reagent/Chemical | Function | Typical Working Concentration | Method Applicability |
|---|---|---|---|
| Acetosyringone | Vir gene inducer, enhances T-DNA transfer | 150-200 µM | Both methods [41] [42] |
| Silwet L-77 | Surfactant, reduces surface tension | 0.02-0.1% | Both methods, particularly vacuum infiltration [40] |
| Cysteine | Antioxidant, reduces tissue browning | Component of infiltration solution | Primarily vacuum infiltration [42] |
| Tween 20 | Surfactant, improves solution penetration | 0.05-0.1% | Primarily vacuum infiltration [42] |
| 6-Benzylaminopurine (6-BA) | Cytokinin, promotes in vitro germination | 0.5 mg/L | Seed soaking with embryo-derived seedlings [41] |
| Gibberellic Acid (GA3) | Plant growth regulator, promotes germination | 1.0 mg/L | Seed soaking with embryo-derived seedlings [41] |
| MS (Murashige and Skoog) Medium | Nutrient base for plant tissue culture | Full or half strength | Both methods, particularly seed soaking [41] |
The choice between vacuum infiltration and seed soaking methods depends on multiple factors, including plant species, experimental objectives, and available resources. The following decision framework can guide researchers in selecting the appropriate method:
Seed soaking is particularly advantageous for laboratories with basic equipment, high-throughput applications, and research focusing on early plant development stages. It demonstrates exceptional performance with embryo-derived seedling systems and enables year-round functional studies without seasonal material constraints [41].
Vacuum infiltration offers superior results for whole-plant transformation, particularly in challenging monocot species, and provides more uniform tissue penetration. It is the method of choice when specialized equipment is available and when researching genes involved in seed germination and early developmental processes [42].
Both methods continue to evolve, with ongoing optimization of parameters such as Agrobacterium strain selection, surfactant compositions, and co-cultivation conditions further enhancing their utility for plant functional genomics and molecular breeding programs [41] [40] [42].
In the field of plant functional genomics, validating experimental systems remains a critical challenge. The phytoene desaturase (PDS) gene has emerged as a cornerstone marker for this purpose, providing a visible and measurable phenotype—photobleaching—that enables researchers to rapidly confirm the efficacy of gene silencing, genome editing, and viral vector systems. As a key enzyme in the carotenoid biosynthesis pathway, PDS catalyzes the conversion of phytoene to ζ-carotene [12]. Its disruption halts carotenoid production, leading to chlorophyll photo-oxidation and the characteristic albino or photobleached appearance in affected tissues [43] [44]. This easily recognizable phenotype makes PDS an invaluable visual reporter for system validation across multiple plant species and technologies, from virus-induced gene silencing (VIGS) to CRISPR/Cas9 genome editing [45] [43] [35].
The utility of PDS extends beyond mere visual confirmation. The photobleaching phenotype directly correlates with molecular efficiency metrics, including reduced target gene expression and successful vector delivery. This dual validation capacity—both visual and molecular—positions PDS as an essential component in the plant biotechnologist's toolkit, particularly for establishing reverse genetics platforms in non-model species where stable transformation systems are often unavailable or inefficient [12] [35].
PDS Applications Across Plant Biotechnology Platforms
Comparative Analysis of PDS Utilization in Key Technologies
Table 1: Applications of PDS as a visual marker across major plant biotechnologies
| Technology | Primary Function | PDS Role | Key Experimental Readouts | Representative Species |
|---|---|---|---|---|
| VIGS [12] [25] [35] | Transient gene silencing | System validation marker | Photobleaching intensity, silencing efficiency (%) | Luffa, Arabidopsis, Atriplex, Tomato |
| CRISPR/Cas9 [43] | Targeted genome editing | Proof-of-concept target | Albino phenotype, mutation efficiency (%) | Chilli pepper, Banana |
| Transcriptional Gene Silencing (TGS) [46] | Epigenetic silencing | Promoter methylation target | Photobleaching strength, methylation percentage | Cotton |
| Genetic Transformation [21] | Stable gene integration | Selection marker | Dwarfism, albinism | Various horticultural species |
Quantitative Performance of PDS Across Experimental Systems
Table 2: Experimental efficiency metrics for PDS-based validation across studies
| Study System | Technology Used | Silencing/Editing Efficiency | Phenotype Observation Timeline | Molecular Validation |
|---|---|---|---|---|
| Luffa acutangula [12] | CGMMV-VIGS | Significant reduction in PDS expression | Not specified | RT-qPCR showing substantial expression reduction |
| Atriplex canescens [35] | TRV-VIGS | 40-80% transcript reduction | ~15 days post-inoculation | qRT-PCR confirmed knockdown |
| Chilli pepper [43] | CRISPR/Cas9 | 62.5% transformed plants edited | Visible in regenerating cotyledons | T7E1 assay, sequence analysis |
| Arabidopsis thaliana [25] | TCV-VIGS | Robust silencing with photobleaching | In systemically infected leaves | Phenotypic observation |
| Iris japonica [45] | TRV-VIGS | 36.67% silencing efficiency | Not specified | Real-time PCR expression reduction |
Experimental Protocols for PDS-Based System Validation
VIGS Implementation Using PDS
The establishment of a Virus-Induced Gene Silencing system typically begins with cloning a fragment of the PDS gene into an appropriate viral vector. For TRV-based systems in species like Atriplex canescens, this involves:
- Gene Fragment Selection: Identify and amplify a 300-400bp conserved region of PDS using tools like SGN-VIGS for optimal fragment selection [35].
- Vector Construction: Clone the PDS fragment into pTRV2 vectors using restriction enzymes (EcoRI and BamHI) or homologous recombination [12] [35].
- Agrobacterium Preparation: Transform constructs into Agrobacterium strains (e.g., GV3101) and culture in YEP medium with appropriate antibiotics until OD600 reaches 0.6-0.8 [12] [35].
- Plant Inoculation: For Atriplex canescens, the optimal method involves vacuum-assisted agroinfiltration (0.5 kPa, 10 min) of germinated seeds with Agrobacterium suspension (OD600 = 0.8) [35]. For Luffa, agroinfiltration is performed on seedlings with two true leaves using a needleless syringe [12].
- Phenotype Monitoring: Systemic photobleaching typically appears in newly emerged leaves approximately 15 days post-inoculation [35].
- Molecular Validation: Confirm silencing efficiency through qRT-PCR analysis of PDS transcript levels in photobleached tissues [12] [35].
CRISPR/Cas9 Genome Editing Using PDS
For CRISPR/Cas9 system validation in chilli pepper, researchers have implemented the following protocol:
- gRNA Design: Identify target sequences in exons 14 and 15 of CaPDS using CHOPCHOP tool, considering GC content, RNA folding, and potential off-target effects [43].
- Vector Construction: Clone two distinct gRNAs targeting CaPDS into a binary vector (e.g., pKSE401) containing Cas9 under 2x CaMV 35S promoter [43].
- Biolistic Transformation: Precipitate plasmid DNA onto gold particles (0.6 μm) and bombard cotyledon explants using PDS-1000/He Gun with parameters: 9 cm flight distance, 900 psi pressure [43].
- Regeneration and Selection: Culture bombarded explants on kanamycin-containing medium to select transformed tissues [43].
- Mutant Screening: Identify successfully edited plants through visible albino phenotypes in cotyledons and confirm with T7 endonuclease assay (T7E1) and sequencing [43].
- Pigment Analysis: Quantify reduction in chlorophyll a, chlorophyll b, and carotenoid content to validate functional disruption of PDS [43].
Diagram 1: Generalized workflow for validating plant biotechnology systems using PDS photobleaching as a visual marker. The process begins with technology selection and proceeds through molecular cloning, delivery, phenotypic screening, and molecular validation.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key research reagents and materials for PDS-based validation experiments
| Reagent/Resource | Function/Purpose | Examples/Specifications |
|---|---|---|
| VIGS Vectors [12] [25] [35] | RNA virus-based delivery of silencing constructs | TRV (Tobacco Rattle Virus), CGMMV (Cucumber Green Mottle Mosaic Virus), TCV (Turnip Crinkle Virus) |
| CRISPR/Cas9 System [43] | Targeted genome editing | pKSE401 vector, Cas9 nuclease, PDS-specific gRNAs |
| Agrobacterium Strains [12] [35] | Delivery of genetic constructs into plant tissues | GV3101, K599 with appropriate antibiotic resistance |
| Online Design Tools [25] [35] | Bioinformatics support for experimental design | SGN-VIGS (fragment selection), CHOPCHOP (gRNA design), Genscript siRNA finder |
| Plant Growth Facilities [12] [35] | Controlled environment for plant growth and phenotype observation | Growth chambers (22-28°C, 16h light/8h dark photoperiod) |
| Molecular Validation Kits [12] [43] | Confirm silencing/editing at molecular level | qRT-PCR reagents, T7E1 assay kits, sequencing services |
Biochemical Pathways and Molecular Mechanisms
Diagram 2: Biochemical pathway of carotenoid biosynthesis showing PDS inhibition point. PDS catalyzes the conversion of phytoene to ζ-carotene; when disrupted, carotenoid production halts, leading to chlorophyll degradation and photobleaching.
The molecular mechanism behind PDS photobleaching involves the carotenoid biosynthesis pathway in plants. PDS performs the second step in this pathway, converting phytoene to ζ-carotene [44]. When PDS is silenced or knocked out through VIGS or CRISPR/Cas9, this conversion is blocked, resulting in:
- Accumulation of phytoene, the colorless substrate of PDS [47]
- Depletion of colored carotenoids including lycopene and downstream products
- Loss of photoprotection for chlorophyll molecules, making them susceptible to photo-oxidation
- Visible photobleaching as chlorophyll degrades, revealing the underlying white tissue
This pathway explains why PDS disruption produces such distinctive phenotypes—the block occurs early enough in the pathway to prevent synthesis of all colored carotenoids, while still allowing accumulation of the colorless phytoene [47] [44]. The resulting loss of photoprotection leads to chlorophyll degradation under light conditions, creating the characteristic white or albino appearance used for visual validation [43].
The phytoene desaturase gene continues to serve as a critical validation marker across plant biotechnology platforms. Its value derives from the easily scorable phenotype of photobleaching, which provides immediate visual confirmation of system functionality without requiring complex instrumentation or destructive sampling [12] [43] [35]. The conservation of PDS across plant species further enhances its utility, enabling protocol transfer between related species and facilitating the establishment of reverse genetics systems in non-model plants [45] [35].
As plant biotechnology advances toward more complex applications—including multiplex gene silencing [25], tissue-specific editing, and epigenetic modification [46]—the fundamental role of PDS as a rapid validation tool becomes increasingly important. The quantitative data and experimental protocols compiled in this guide provide researchers with a comprehensive resource for implementing PDS-based validation in their systems, accelerating functional genomics research across diverse plant species.
Virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics tool for rapidly characterizing gene functions in plants, particularly in species where stable genetic transformation remains challenging. This technology leverages the plant's innate RNA-based antiviral defense system to achieve sequence-specific downregulation of target genes, enabling functional genomics studies without the need for stable transformation. The application of VIGS has expanded beyond model plants to encompass numerous horticultural crops, providing invaluable insights into gene functions underlying agronomically important traits. This review presents a comparative analysis of VIGS implementation in three economically significant crops—walnut, luffa, and watermelon—highlighting the methodological adaptations required for different plant systems and their contributions to validating plant gene function within the broader context of functional genomics research.
Comparative Analysis of VIGS Applications
Table 1: Comparative overview of VIGS implementation across three crop species
| Crop Species | Viral Vector | Target Genes | Key Optimization Parameters | Silencing Efficiency | Primary Applications |
|---|---|---|---|---|---|
| Walnut (Juglans regia L.) | Tobacco Rattle Virus (TRV) | Phytoene desaturase (JrPDS) | Infiltration method, fragment length (255 bp), Agrobacterium density (OD~600~=0.8) | Up to 48% | Functional genomics in woody plants, abiotic stress research [48] |
| Ridge Gourd (Luffa acutangula) | Cucumber Green Mottle Mosaic Virus (CGMMV) | LaPDS, Tendril synthesis (LaTEN) | Agroinfiltration at two true-leaf stage, OD~600~=0.8-1.0 | Significant reduction in gene expression | Study of developmental traits, rapid gene function validation [4] |
| Watermelon (Citrullus lanatus) | CGMMV, CFMMV | PDS, male sterility genes | Not specified in available data | Effective silencing with phenotypic consequences | Fruit quality improvement, reproductive development studies [4] [3] |
Table 2: Phenotypic consequences of successful VIGS in target crops
| Crop Species | Target Gene | Observed Silencing Phenotype | Validation Method | Functional Significance |
|---|---|---|---|---|
| Walnut | JrPDS | Photobleaching in leaves | Visible bleaching, RT-qPCR | Carotenoid biosynthesis marker [48] |
| Ridge Gourd | LaPDS | Photobleaching in leaves and stems | Visible bleaching, RT-qPCR | Carotenoid biosynthesis marker [4] |
| Ridge Gourd | LaTEN | Shorter tendril length, higher nodal positions for tendrils | Morphometric analysis, RT-qPCR | Tendril development regulation [4] |
| Watermelon | PDS | Photobleaching | Visible bleaching | Carotenoid biosynthesis marker [4] [3] |
| Watermelon | Male sterility genes | Abnormal stamens, no pollen | Morphological analysis | Reproductive development [4] |
VIGS Workflow and Mechanism
The fundamental VIGS mechanism exploits the plant's post-transcriptional gene silencing (PTGS) machinery, which normally functions as an antiviral defense system. When plants encounter viral infection, they recognize and process double-stranded RNA intermediates of viral replication through Dicer-like enzymes, generating 21-24 nucleotide small interfering RNAs (siRNAs). These siRNAs are incorporated into the RNA-induced silencing complex (RISC), which guides sequence-specific degradation of complementary viral RNA sequences. In VIGS, this system is co-opted by engineering viral vectors to carry fragments of host plant genes, leading to degradation of corresponding endogenous mRNAs and thus knockdown of target gene expression [4] [3].
The following diagram illustrates the generalized VIGS workflow and molecular mechanism:
Detailed Case Studies
VIGS in Walnut (Juglans regia L.)
Experimental Protocol: The establishment of an efficient TRV-based VIGS system in walnut addressed a significant methodological gap for this economically important tree species. Researchers optimized multiple parameters to achieve effective silencing:
- Plant Material: Germinated seeds of 'Qingxiang' and 'Xiangling' walnut cultivars were grown under controlled conditions (24°C, 16h/8h light/dark cycle) [48].
- Vector Construction: A 255bp fragment of the walnut JrPDS gene (LOC108996675) was cloned into the TRV2 vector, while TRV1 contained genes for viral replication and movement [48].
- Agrobacterium Preparation: GV3101 strains carrying pTRV1 and pTRV2-derived vectors were cultured to OD~600~=0.8, harvested, and resuspended in infiltration buffer (10 mM MgCl~2~, 10 mM MES, 200 μM AS) [48].
- Infiltration Methods: Three infiltration techniques were evaluated, with needleless syringe infiltration proving most effective for walnut seedlings [48].
- Validation: Silencing efficiency was assessed through visible photobleaching phenotypes and confirmed by RT-qPCR analysis showing significant reduction in JrPDS transcript levels [48].
This optimized protocol achieved up to 48% silencing efficiency, representing a breakthrough for functional genomics in walnut, where traditional transformation systems remain challenging [48].
VIGS in Ridge Gourd (Luffa acutangula)
Experimental Protocol: The CGMMV-based VIGS system established for ridge gourd enables rapid functional analysis of genes involved in development and metabolism:
- Vector System: The pV190 CGMMV vector was used to construct recombinant vectors carrying fragments of LaPDS (marker gene) and LaTEN (tendril development gene) [4].
- Plant Inoculation: Agrobacterium tumefaciens GV3101 harboring the VIGS constructs was inoculated into two-leaf-stage seedlings through agroinfiltration with needleless syringes [4].
- Optimized Conditions: Bacterial suspensions were adjusted to OD~600~=0.8-1.0 and maintained at room temperature for 2+ hours before inoculation [4].
- Post-Inoculation Care: Inoculated plants were kept under high humidity for 24 hours before transition to normal growth conditions (28°C/24°C, 16h/8h light/dark) [4].
Functional Validation: Beyond the marker LaPDS gene, which showed characteristic photobleaching, researchers targeted LaTEN, encoding a CYC/TB1-like transcription factor involved in tendril development. Plants silenced for LaTEN exhibited distinct morphological changes—shorter tendrils and altered nodal positions for tendril emergence—demonstrating the utility of this system for studying developmental genes. RT-qPCR confirmed significant reduction in target gene expression, validating the system's efficiency [4].
VIGS in Watermelon (Citrullus lanatus)
Implementation and Applications: While detailed methodological protocols for watermelon were not provided in the available literature, successful VIGS applications in watermelon have been reported using both CGMMV and Cucumber Fruit Mottle Mosaic Virus (CFMMV) vectors [4] [3]. The CGMMV-based system has been effectively used to silence the PDS gene, producing the characteristic photobleaching phenotype [4]. More significantly, CFMMV-based VIGS has enabled functional studies of genes related to male sterility, with silencing of 8 out of 38 candidate genes resulting in abnormal stamen development and complete pollen absence [4]. These applications highlight the value of VIGS for studying reproductive traits and facilitating watermelon breeding programs.
Essential Research Reagent Solutions
Table 3: Key research reagents and materials for VIGS implementation
| Reagent/Material | Function in VIGS | Examples from Case Studies | Specific Application Notes |
|---|---|---|---|
| Viral Vectors | Delivery of target gene fragments | TRV, CGMMV, CFMMV | TRV: Broad host range, meristem penetration; CGMMV: Cucurbit-specific [4] [3] [48] |
| Agrobacterium Strains | Mediate plant transformation | GV3101 | Standard laboratory workhorse for VIGS delivery [4] [48] [49] |
| Marker Genes | System validation | PDS, ChlH | Visible photobleaching phenotypes confirm silencing efficiency [4] [48] [49] |
| Infiltration Buffers | Agrobacterium suspension medium | 10 mM MgCl~2~, 10 mM MES, 200 μM AS | Maintains bacterial viability during inoculation [4] [48] |
| Antibiotics | Selection pressure for vectors | Kanamycin, Rifampicin | Maintain plasmid integrity in bacterial cultures [4] [48] |
| PCR Components | Target fragment amplification | Phanta Max Super-Fidelity DNA Polymerase | High-fidelity amplification of gene fragments for cloning [4] |
Discussion and Future Perspectives
The case studies presented herein demonstrate that VIGS technology has evolved into a versatile platform for functional genomics across diverse plant species, each requiring specific adaptations. The successful implementation in walnut, a woody species with notoriously difficult transformation systems, underscores the particular value of VIGS for recalcitrant species [48]. Similarly, the application in luffa provides a rapid alternative to conventional transformation methods that have proven challenging in cucurbits [4]. These advances are particularly timely given the expanding genomic resources for many crop species, which have created an urgent need for efficient functional validation tools.
Future developments in VIGS technology will likely focus on several key areas: (1) expanding the host range of viral vectors to encompass more crop species, (2) improving silencing efficiency and persistence, particularly in meristematic tissues, (3) developing inducible and tissue-specific systems for precise spatiotemporal control of silencing, and (4) integrating VIGS with emerging technologies such as CRISPR/Cas for comprehensive functional genomics platforms [3]. The combination of VIGS with multi-omics approaches will further accelerate the discovery and validation of genes controlling agronomically important traits, ultimately enhancing crop improvement programs.
VIGS has transformed functional genomics in plants by providing a rapid, cost-effective alternative to stable transformation for gene function characterization. The successful implementation of VIGS in walnut, luffa, and watermelon—each with distinct biological characteristics and methodological requirements—demonstrates the remarkable adaptability of this technology. As genomic resources continue to expand for non-model crops, VIGS will play an increasingly vital role in bridging the gap between gene sequence information and biological function, ultimately accelerating crop improvement efforts and advancing our fundamental understanding of plant biology.
High-Throughput Functional Screening for Male Sterility and Stress Resistance Genes
The completion of genome sequencing for numerous plant species has generated vast amounts of genomic data, creating an urgent need for efficient functional characterization of genes. While powerful genome-editing techniques like CRISPR/Cas9 exist for functional characterization, they are often labor-intensive, costly, and reliant on stable transformation, which is particularly challenging for recalcitrant species. Virus-induced gene silencing (VIGS) has emerged as a potent and flexible alternative that bypasses these limitations, enabling high-throughput functional screening without the need for stable transformation [3] [50].
The application of high-throughput functional screening is particularly valuable for studying complex agronomic traits such as male sterility and stress resistance. Male sterility, characterized by the inability to produce functional pollen while maintaining female fertility, provides crucial breeding tools for harnessing heterosis in crop species. Similarly, understanding the genetic architecture of stress resistance requires systematic analysis of numerous candidate genes. This guide compares the leading technologies for high-throughput functional screening in plants, with a special emphasis on VIGS applications for male sterility and stress resistance genes, providing researchers with experimental frameworks and comparative data to inform their study designs [51] [52].
Technology Comparison: VIGS Versus Alternative Functional Screening Methods
Table 1: Comparative analysis of major technologies for high-throughput functional gene screening in plants
| Technology | Key Features | Throughput Capacity | Time Requirements | Key Advantages | Major Limitations |
|---|---|---|---|---|---|
| VIGS | Transient, sequence-specific post-transcriptional gene silencing | High (can screen dozens of genes simultaneously) | 3-8 weeks (from inoculation to phenotype) | No stable transformation required; applicable to non-model species; cost-effective | Transient effect; potential viral symptoms; variable silencing efficiency |
| CRISPR/Cas9 | Permanent gene editing via DNA double-strand breaks | Moderate to high (depends on transformation efficiency) | 6-12 months (for stable lines) | Permanent genetic changes; precise editing; versatile applications | Requires stable transformation; time-consuming; off-target effects possible |
| Chemical Genetics | Small molecule-induced phenotypic changes | Very high (can screen thousands of compounds) | Days to weeks (rapid phenotype assessment) | Conditional and reversible; dosage-dependent; no genetic modification required | Target identification challenging; potential non-specific effects |
| TILLING/ Mutagenesis | Identification of mutations in target genes | Moderate (depends on population size) | 12-24 months (for population development and screening) | Non-GMO approach; generates allelic series; applicable to any species | Background mutations; laborious screening; not targeted for specific genes |
VIGS Systems and Vector Selection for High-Throughput Screening
VIGS technology leverages the plant's natural RNA silencing machinery as a defense mechanism against viral infections. When recombinant viral vectors carrying host gene fragments infect plants, the resulting sequence-specific RNA degradation targets both viral RNA and homologous endogenous mRNAs for post-transcriptional silencing. The effectiveness of VIGS for high-throughput screening depends on selecting appropriate viral vectors optimized for specific plant families [3] [4].
Table 2: Major VIGS vectors and their applications in male sterility and stress resistance research
| Vector System | Virus Type | Host Range | Key Applications in Literature | Silencing Efficiency | Notable Features |
|---|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) | RNA virus | Broad (especially Solanaceae) | Pepper fruit development, disease resistance genes | High | Efficient systemic movement; targets meristematic tissues |
| Cucumber Fruit Mottle Mosaic Virus (CFMMV) | Tobamovirus | Cucurbits (watermelon, melon, cucumber) | Male sterility genes in watermelon | High in cucurbits | Monopartite genome; relatively easy to manipulate |
| Barley Stripe Mosaic Virus (BSMV) | RNA virus | Monocots (wheat, barley, Brachypodium) | Stress resistance genes in wheat | Moderate to high | Effective in cereal crops and roots |
| Cucumber Green Mottle Mosaic Virus (CGMMV) | Tobamovirus | Cucurbits (Luffa, cucumber, watermelon) | Tendril development in Luffa | High in cucurbits | Recently established for Luffa species |
The TRV-based system remains one of the most versatile and widely used VIGS systems, particularly for Solanaceae plants like pepper and tomato. Its bipartite genome organization requires two vectors: TRV1, encoding replicase and movement proteins, and TRV2, containing the capsid protein gene and a multiple cloning site for inserting target sequences. For cucurbit species, which are generally recalcitrant to genetic transformation, CFMMV-based vectors have demonstrated remarkable efficiency, successfully silencing eight out of 38 candidate male sterility genes in watermelon, producing sterile male flowers with abnormal stamens and no pollen [3] [50].
Experimental Design and Workflow for High-Throughput VIGS Screening
The successful implementation of VIGS for high-throughput screening requires careful optimization of multiple parameters. Key factors influencing silencing efficiency include insert design, agroinfiltration methodology, plant developmental stage, agroinoculum concentration, plant genotype, and environmental conditions such as temperature, humidity, and photoperiod [3].
Diagram 1: High-throughput VIGS screening workflow highlighting key experimental phases. Critical optimization steps are shown in yellow, while validation phases are in green.
Protocol: CFMMV-Based VIGS for Male Sterility Gene Screening
The following detailed protocol has been successfully implemented for high-throughput screening of male sterility genes in watermelon, demonstrating the potential of VIGS for functional genomics in transformation-recalcitrant species [50]:
Insert Fragment Selection and Cloning:
- Amplify 200-300 bp fragments of candidate genes using Phanta Max Super-Fidelity DNA Polymerase
- Design primers with appropriate restriction sites (e.g., BamHI for pCF93 vector system)
- Clone fragments into CFMMV-based vector (pCF93) using homologous recombinase
- Transform into E. coli DH5α and verify inserts by colony PCR and sequencing
Agrobacterium Preparation:
- Transform recombinant plasmid into Agrobacterium tumefaciens GV3101
- Culture single colonies in YEP liquid medium with appropriate antibiotics (50 mg/L Kan, 25 mg/L Rif)
- Resuspend bacterial pellets in infiltration buffer (10 mM MgCl₂, 10 mM MES, 200 μM AS)
- Adjust suspension to OD₆₀₀ of 0.8-1.0 and incubate at room temperature for 2+ hours
Plant Inoculation:
- Use seedlings at two-true-leaf stage for agroinfiltration
- Gently create small holes on cotyledons and true leaves using syringe needle
- Infiltrate bacterial suspension from abaxial side of leaves with needleless syringe
- Maintain high humidity for 24 hours post-inoculation, then normal growth conditions
Phenotypic Assessment:
- Monitor plants for development of male-sterile phenotypes (abnormal stamens, no pollen)
- Document phenotypic changes with photographic evidence
- Collect tissue samples for molecular validation of silencing efficiency
This protocol enabled researchers to characterize the function of 38 candidate male sterility genes simultaneously in watermelon, identifying 8 genes that produced male-sterile phenotypes when silenced [50].
Case Studies: Applications in Male Sterility and Stress Resistance Research
Male Sterility Gene Discovery
Male sterility can be classified as either cytoplasmic male sterility (CMS), involving mitochondrial-nuclear genome interactions, or genic male sterility (GMS), caused by nuclear genes alone. High-throughput VIGS screening has accelerated the identification of genes involved in both pathways by enabling rapid functional testing of candidates identified from transcriptomic and proteomic studies [51] [52].
In a landmark study, researchers combined transcriptome and proteome sequencing of a thermo-sensitive male sterile wheat line (KTM3315A) under different fertility conditions using RNA-seq and iTRAQ techniques. This approach identified TaEXPB5, a gene encoding an expansin protein, as critical for pollen development. The function was validated using BSMV-based VIGS in wheat, confirming that silencing TaEXPB5 led to abnormal pollen development and male sterility. This study demonstrated the power of integrating multi-omics data with VIGS for rapid gene function characterization [53].
Non-coding RNAs have also been identified as key regulators of male sterility. MicroRNAs including miR160, miR167, and miR157 interact with auxin response factors (ARFs) and participate in hormone signaling pathways that regulate stamen development, tapetum degradation, microspore formation, and pollen release. VIGS provides an efficient tool for functional characterization of these regulatory networks in species where stable transformation is challenging [51].
Stress Resistance Gene Discovery
High-throughput phenotyping platforms have revolutionized stress resistance gene discovery by enabling non-destructive monitoring of plant responses to abiotic stresses over time. These systems employ multiple sensing technologies including RGB imaging, hyperspectral imaging (HSI), and X-ray computed tomography (CT) to capture both external and internal plant responses to stress conditions [54].
In a comprehensive study of maize drought tolerance, researchers used high-throughput multiple optical phenotyping to monitor 368 maize genotypes under drought stress over 98 days. The platform extracted 10,080 effective image-based traits (i-traits) that served as indicators of maize drought responses. Combining this high-throughput phenotyping with genome-wide association studies (GWAS) revealed 4,322 significant locus-trait associations, representing 1,529 quantitative trait loci and 2,318 candidate genes. The researchers validated two novel genes, ZmcPGM2 and ZmFAB1A, which regulate i-traits and drought tolerance, demonstrating the power of this integrated approach [54].
Diagram 2: Integrated workflow for stress resistance gene discovery combining high-throughput phenotyping, GWAS, and VIGS validation.
Advanced Applications and Integration with Novel Technologies
Machine Learning-Enhanced Phenotype Analysis
The enormous volume, variety, and velocity of data generated by high-throughput phenotyping platforms creates a 'big data' challenge that requires sophisticated analytical approaches. Machine learning algorithms have emerged as powerful tools for faster, more efficient, and better data analytics in plant phenotyping [55].
Convolutional neural networks (CNNs) have been successfully applied to classify plant growth responses in high-throughput chemical genetic screens. In one study, researchers employed residual neural network (ResNet) architecture to classify images of Arabidopsis seedlings as either normal or altered growth with 100% accuracy, enabling efficient screening of chemical libraries for genotype-specific effects. These deep learning approaches automatically extract discriminative features from raw image data and rapidly predict large datasets, making them particularly valuable for large-scale screening projects [56].
Integration with Genome Editing
While VIGS remains invaluable for initial high-throughput screening, CRISPR/Cas9 genome editing provides complementary technology for detailed functional validation. The most effective research pipelines now use VIGS for primary screening of multiple candidate genes followed by CRISPR/Cas9 for precise manipulation of the most promising targets [52].
This integrated approach is particularly valuable for male sterility research, where genome editing can create stable sterile lines for hybrid breeding programs. Recent advances have enabled the development of transgenerational gene editing systems that introduce desirable alleles without permanent transgenic elements, addressing regulatory concerns and facilitating commercial application [52].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key research reagents and materials for high-throughput functional screening experiments
| Reagent/Resource | Specifications | Application | Example Uses |
|---|---|---|---|
| VIGS Vectors | TRV, CFMMV, BSMV, CGMMV with multiple cloning sites | Target gene silencing in specific plant families | Silencing PDS as visual marker; testing candidate gene function |
| Agrobacterium Strains | GV3101, EHA105, LBA4404 with appropriate virulence | Delivery of VIGS constructs into plant tissues | Agroinfiltration of seedlings; vacuum infiltration |
| Infiltration Buffers | 10 mM MgCl₂, 10 mM MES, 200 μM acetosyringone | Enhancement of T-DNA transfer efficiency | Preparation of Agrobacterium suspension for inoculation |
| High-Throughput Phenotyping Systems | RGB, hyperspectral, and X-ray CT imaging capabilities | Non-destructive monitoring of plant growth and development | Dynamic assessment of stress responses; quantification of morphological traits |
| Reference Genes | EF1α, ACTIN, UBQ with stable expression across conditions | qRT-PCR normalization for silencing efficiency verification | Molecular validation of target gene knockdown in VIGS experiments |
High-throughput functional screening technologies, particularly VIGS, have dramatically accelerated the pace of gene function characterization in plants. The integration of these tools with advanced phenotyping platforms, multi-omics approaches, and machine learning analytics has created powerful pipelines for dissecting complex biological processes including male sterility and stress resistance.
Future developments in this field will likely focus on increasing screening throughput, improving silencing efficiency in recalcitrant species, and enhancing spatial-temporal control of gene silencing. The combination of VIGS with single-cell transcriptomics and proteomics promises to elucidate cell-type-specific gene functions, while advances in virus-mediated gene editing may enable high-throughput knockout screening. As these technologies continue to mature, they will play an increasingly important role in accelerating crop improvement programs and addressing global food security challenges.
Enhancing Silencing Efficiency: Critical Factors and Solutions
Optimizing Agrobacterium Concentration (OD600) and Infiltration Conditions
Publish Comparison Guides
In plant functional genomics, the validation of gene function through techniques like Virus-Induced Gene Silencing (VIGS) and knockout studies fundamentally relies on efficient transgene delivery. Agrobacterium-mediated transformation serves as a cornerstone for these investigations, with the optical density at 600 nm (OD600) of the bacterial suspension representing a critical parameter that directly influences transformation efficiency and experimental outcomes. Suboptimal OD600 can lead to either insufficient T-DNA delivery or excessive bacterial stress, both compromising results. This guide provides a systematic comparison of optimized OD600 values and complementary infiltration conditions across diverse plant species and transformation methods, offering researchers a consolidated resource for experimental design. The data presented herein enables scientists to select appropriate parameters for their specific plant system, thereby enhancing the reliability of gene function validation within the broader context of plant molecular biology research.
Comparative Analysis of Optimal Agrobacterium Concentration (OD600) Across Methods
Table 1: Comparative analysis of optimal Agrobacterium concentration (OD600) across transformation methods and plant species.
| Transformation Method | Plant Species | Optimal OD600 | Key Supporting Conditions | Reported Efficiency/Outcome | Primary Citation |
|---|---|---|---|---|---|
| Syringe Infiltration (Transient) | Sunflower (Helianthus annuus) | 0.8 | 0.02% Silwet L-77 surfactant | ~90% transformation efficiency [40] | |
| Seedling Infiltration (Transient) | Sunflower (Helianthus annuus) | 0.8 | 0.02% Silwet L-77, 2-hour immersion | ~90% transformation efficiency [40] | |
| Hairy Root Transformation | Woodland Strawberry (Fragaria vesca) | 0.7 | 10-minute infection, 4-day co-cultivation | Up to 71.43% transformation efficiency [57] | |
| Hairy Root Transformation | Passion Fruit (Passiflora edulis) | 0.6 | 100 µM acetosyringone, 30-minute infection, 2-day dark co-cultivation | 11.3% transformation efficiency with strain K599 [58] | |
| VIGS Assay | Ridge Gourd (Luffa acutangula) | 0.8 - 1.0 | 200 µM acetosyringone in resuspension buffer | Effective silencing of PDS and TEN genes [12] | |
| Simplified Transient Assay | Nicotiana benthamiana | ~0.1 - 1.0 (visual estimate) | 100 µM acetosyringone, co-infiltration with p19 silencing suppressor | Visible cell death phenotype with rice XB3 gene [59] |
Detailed Experimental Protocols for Key Methods
Agrobacterium-Mediated Syringe Infiltration in Sunflower
The optimized protocol for sunflower transient transformation achieves high efficiency through specific parameters for syringe infiltration and whole-seedling immersion [40].
- Agrobacterium Preparation: Use Agrobacterium tumefaciens strain GV3101 harboring the binary vector of interest (e.g., pBI121 with a GUS reporter). Grow the primary culture in LB with appropriate antibiotics to saturation. Dilute the culture to an OD600 of 0.8 in an infiltration medium. The addition of 0.02% Silwet L-77 as a surfactant is crucial for promoting bacterial entry into plant tissues [40].
- Plant Material: Utilize sunflower seedlings grown hydroponically for 3 days for the infiltration method, or seedlings grown in soil for 4-6 days for the injection method.
- Infiltration Procedure:
- For syringe infiltration, gently press a needleless syringe containing the bacterial suspension against the abaxial side of a cotyledon and apply steady pressure to infiltrate the tissue. The area should darken as the suspension fills the intercellular spaces.
- For whole-seedling infiltration, immerse the entire seedling in the bacterial suspension for 2 hours with gentle agitation.
- Post-Infiltration Handling: After infiltration, culture the plants in the dark at room temperature for three days to allow for transgene expression before analysis. GUS staining or molecular analysis can confirm transformation success.
Hairy Root Transformation in Woodland Strawberry
This protocol for strawberry hairy root transformation highlights the importance of explant type and co-cultivation duration for high efficiency [57].
- Agrobacterium Preparation: Use A. rhizogenes strains MSU440 or C58C1 carrying the desired binary vector (e.g., 35S::eGFP). Grow the bacteria to an OD600 of 0.7 in a suitable medium.
- Explants: Hypocotyls from 7-day-old in vitro-grown strawberry seedlings (YW5AF7 genotype) are the most effective explants.
- Inoculation and Co-cultivation: Immerse the hypocotyl explants in the bacterial suspension for 10 minutes. Blot the explants dry and transfer them to co-cultivation media. Co-cultivate for four days in the dark.
- Hairy Root Induction and Selection: After co-cultivation, transfer the explants to a selective medium (e.g., containing antibiotics to counter Agrobacterium and select for transformed roots). Transgenic hairy roots, which are highly branched and can be identified by fluorescence if a reporter like eGFP is used, typically emerge within 2-3 weeks.
VIGS Assay in Ridge Gourd
This protocol uses a CGMMV-based vector for virus-induced gene silencing in the cucurbit species, ridge gourd [12].
- Vector and Agrobacterium Preparation: Use the pV190 vector (a CGMMV-based VIGS vector) harboring a fragment of the target gene (e.g., Phytoene desaturase (PDS)). Transform the vector into A. tumefaciens strain GV3101.
- Culture Preparation: Grow the agrobacterial culture to an OD600 between 0.8 and 1.0. Pellet the bacteria and resuspend them in an induction buffer (10 mM MgCl₂, 10 mM MES, and 200 µM acetosyringone). Allow the suspension to incubate at room temperature for over 2 hours to activate virulence genes.
- Plant Infiltration: Use ridge gourd seedlings at the two-true-leaf stage. Gently puncture the cotyledons or true leaves with a needle. Using a needleless syringe, infiltrate the bacterial suspension from the abaxial (lower) side of the leaf.
- Post-Infiltration Care: Maintain the plants under high humidity for 1-2 days after infiltration. Silencing phenotypes, such as photobleaching for PDS, can be observed within a few weeks. Silencing efficiency should be validated using RT-qPCR to measure the reduction in endogenous target gene expression.
Workflow for Parameter Optimization
The following diagram illustrates a generalized decision-making workflow for optimizing Agrobacterium concentration and infiltration conditions, based on the experimental goals and plant material.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key reagents and materials for Agrobacterium-mediated transformation protocols.
| Reagent/Material | Function/Role | Example Usage & Concentration |
|---|---|---|
| Silwet L-77 | Surfactant that reduces surface tension, enabling the bacterial suspension to spread and infiltrate plant tissues effectively. | Used at 0.02% in sunflower infiltration and injection methods [40]. |
| Acetosyringone (AS) | A phenolic compound that activates the Agrobacterium Vir genes, which are essential for T-DNA transfer into the plant cell. | Used at 200 µM in VIGS assays for ridge gourd [12] and 100 µM in passion fruit hairy root transformation [58]. |
| MgCl₂ and MES Buffer | Components of the infiltration buffer; provide a suitable ionic environment and pH stability for the Agrobacterium during the infection process. | Standard infiltration buffer: 10 mM MgCl₂, 10 mM MES, pH 5.6-5.7 [12] [59]. |
| p19 Silencing Suppressor | A viral protein that suppresses the plant's post-transcriptional gene silencing (PTGS) machinery, thereby boosting transient expression levels of the transgene. | Co-infiltrated with the gene of interest in N. benthamiana to achieve high-level protein expression [59]. |
| Strain-Specific Antibiotics | Selective agents (e.g., Kanamycin, Rifampicin) added to culture media to maintain the binary vector and ensure the purity of the Agrobacterium strain. | Commonly used in primary and secondary culture growth (e.g., 50-100 µg/mL Kanamycin) [59]. |
This guide has systematically compared optimal Agrobacterium concentrations and infiltration conditions across a spectrum of plant species and transformation methodologies. The data unequivocally demonstrates that while an OD600 of 0.6 to 1.0 is generally effective, the precise optimum is contingent upon the specific method, plant species, and Agrobacterium strain employed. The integration of optimized supporting factors—such as the use of surfactants like Silwet L-77, virulence inducers like acetosyringone, and gene silencing suppressors like p19—is equally critical for achieving high efficiency.
For researchers engaged in validating plant gene function via VIGS or knockout studies, these findings underscore the necessity of a tailored, systematic approach. Beginning with the established parameters for a related species and conducting pilot experiments to fine-tune the OD600 and other conditions will yield the most robust and reproducible results, ultimately accelerating the pace of discovery in plant functional genomics.
In the field of plant functional genomics, validating gene function requires robust reverse genetics tools. Among these, Virus-Induced Gene Silencing (VIGS) has emerged as a powerful technique for transient gene knockdown, complementing permanent knockout studies. VIGS operates by harnessing the plant's innate RNA-mediated antiviral defense mechanism to degrade target endogenous mRNAs, enabling rapid assessment of gene function without stable transformation. The efficiency of VIGS, however, is not guaranteed—it hinges critically on two fundamental experimental parameters: the size of the inserted gene fragment and the strategic design of the VIGS vector itself. For researchers aiming to maximize silencing efficacy, understanding and optimizing these parameters is essential for generating reliable, reproducible phenotypic data in functional genomics studies.
VIGS Fragment Size: A Quantitative Analysis
The length of the target gene fragment inserted into the VIGS vector is a primary determinant of silencing efficiency. Fragments that are too short may lack specificity, while excessively long inserts can compromise viral replication or movement, reducing systemic silencing. Recent studies across diverse plant species provide quantitative guidance for optimal fragment selection.
Table 1: Optimal VIGS Fragment Sizes Across Plant Species
| Plant Species | Target Gene | Optimal Fragment Size | Silencing Efficiency | Citation |
|---|---|---|---|---|
| Walnut (Juglans regia L.) | JrPOR | 255 bp | Up to 48% | [60] |
| Tea plant (Camellia sinensis L.) | CsPDS | ~300 bp | Up to 63.3% | [61] |
| Atriplex (Atriplex canescens) | AcPDS | 300-400 bp | 40-80% transcript reduction | [35] |
The consensus from these studies indicates that fragment sizes in the 250-400 base pair range typically yield the highest silencing efficiency. Research in walnut demonstrated that a 255 bp fragment was optimal for silencing the JrPOR gene [60]. Similarly, a study in the tea plant Camellia sinensis utilized a fragment of approximately 300 bp to silence the CsPDS gene, achieving a high silencing efficiency of 63.3% [61]. Beyond mere size, sequence specificity is equally critical. Bioinformatics tools, such as the SGN-VIGS online tool, are recommended to predict unique target regions with minimal off-target potential, ensuring that the designed fragment selectively silences the intended gene [35].
VIGS Vector Insert Position and Configuration
The positional considerations for VIGS efficacy operate on two levels: the location of the target fragment within the viral genome and the physical placement of the viral vector within plant tissues.
Vector Engineering and Insert Position
The architecture of the VIGS vector itself can be modified to enhance systemic infection, particularly in challenging tissues like roots. A landmark study demonstrated that a modified Tobacco Rattle Virus (TRV) vector retaining the helper protein 2b—a component often deleted in earlier constructs—dramatically improved root invasion and meristematic tissue infection [7]. This TRV-2b vector exhibited a higher percentage of systemic infection (60% in N. benthamiana, 74% in Arabidopsis) compared to the TRV-Δ2b vector (23% and 30%, respectively), leading to a more pervasive VIGS response in root tissues [7]. This finding highlights that the genetic context of the insert within the viral backbone is a crucial factor for tropism and silencing efficiency.
Tissue-Based Insertion Strategies
The method of vector delivery, or "insertion" into the plant, directly impacts the localization and strength of silencing.
Table 2: VIGS Inoculation Methods and Efficiency
| Inoculation Method | Plant Species | Key Optimized Parameters | Relative Efficiency | Citation |
|---|---|---|---|---|
| Leaf Injection | Walnut (Juglans regia) | Agrobacterium OD600 = 1.1 | More effective than spraying | [60] |
| Vacuum Infiltration | Tea Plant (Camellia sinensis) | 0.8 kPa for 5 minutes | Highest efficiency (63.3%) | [61] |
| Vacuum Infiltration | Atriplex (Atriplex canescens) | 0.5 kPa for 10 min on germinated seeds | ~16.4% plant efficiency | [35] |
For above-ground tissues, leaf injection is a reliable and widely used technique. In walnut seedlings, this method was found to be more effective than spray infiltration for delivering the silencing signal [60]. For more challenging tissues or difficult-to-transform species, vacuum-assisted agroinfiltration has proven superior. By subjecting germinated seeds or whole seedlings to a brief vacuum while submerged in an Agrobacterium suspension containing the VIGS vector, the solution is physically drawn into the intercellular spaces. This method achieved the highest reported silencing efficiency of 63.3% in tea plants [61] and was successfully established for Atriplex canescens [35].
Experimental Protocols for VIGS Optimization
Protocol 1: Determining Optimal Fragment Size
- Gene Fragment Selection: Identify the open reading frame (ORF) of your target gene. Using tools like the SGN-VIGS online predictor, select three non-overlapping fragments of varying lengths (e.g., 250 bp, 300 bp, 400 bp) from the 5' end, center, and 3' end of the gene [35].
- Specificity Check: Verify the specificity of each candidate fragment using Nucleotide-BLAST against the host plant's genome to minimize off-target silencing.
- Vector Construction: Amplify the selected fragments via PCR using primers flanked by appropriate restriction sites (e.g., EcoRI and BamHI). Clone each fragment into the multiple cloning site of a VIGS vector, such as pTRV2 [35].
- Efficiency Testing: Transform the constructed vectors into Agrobacterium and inoculate host plants. Assess silencing efficiency through phenotypic observation (e.g., photobleaching for PDS), quantitative RT-PCR to measure transcript abundance reduction, and calculation of the percentage of plants showing silencing.
Protocol 2: High-Efficiency Vacuum Infiltration
- Plant Material Preparation: Use germinated seeds with radicles of 1-3 cm in length, as this developmental stage shows high susceptibility [35].
- Agrobacterium Preparation: Transform the pTRV1 and pTRV2 (with insert) vectors into Agrobacterium strain GV3101. Grow cultures to mid-log phase (OD600 = 0.6-0.8) and resuspend in an infiltration buffer containing 10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone to an final OD600 of 0.8-1.0 [35].
- Vacuum Infiltration: Mix the pTRV1 and pTRV2 Agrobacterium suspensions in a 1:1 ratio. Submerge the plant materials in the mixture and apply a vacuum of 0.5-0.8 kPa for 5-10 minutes [61] [35].
- Plant Recovery and Analysis: Gently rinse the materials with water, transplant them into growing medium, and maintain under controlled conditions. Monitor for the emergence of systemic silencing phenotypes in new leaves, typically appearing 2-3 weeks post-inoculation.
VIGS Experimental Workflow for Maximum Silencing
The Scientist's Toolkit: Key Research Reagents
A successful VIGS experiment relies on a core set of biological and chemical reagents, each playing a critical role in the process.
Table 3: Essential Research Reagents for VIGS Experiments
| Reagent / Material | Function in VIGS Protocol | Specific Application Example |
|---|---|---|
| pTRV1 & pTRV2 Vectors | Binary TRV vectors for agroinfiltration; pTRV1 contains replication genes, pTRV2 carries the target gene insert. | Standard vector system used in walnut, tea, Atriplex, and Arabidopsis [60] [61] [7]. |
| Agrobacterium tumefaciens (e.g., GV3101) | Delivery vehicle for transferring TRV vectors into plant cells. | Strain used for efficient T-DNA transfer in protocols for walnut and Atriplex [60] [35]. |
| Acetosyringone | Phenolic compound that induces Agrobacterium virulence genes, enhancing T-DNA transfer. | Added to the infiltration buffer in protocols for tea plants and Atriplex [61] [35]. |
| Silwet-77 (Surfactant) | Reduces surface tension of the infiltration buffer, improving leaf wetting and Agrobacterium penetration. | Included in the infiltration buffer for Atriplex canescens [35]. |
| Phytoene Desaturase (PDS) Gene | Endogenous reporter gene; silencing causes visible photobleaching, allowing for visual assessment of VIGS efficiency. | Used as a visual marker to optimize systems in Atriplex, tea, and walnut [60] [61] [35]. |
Within the broader context of plant functional genomics, VIGS stands as an indispensable tool for the initial, high-throughput validation of gene function before committing to the creation of stable knockouts. The journey to maximum silencing efficiency is a deliberate one, guided by precise parameters. The experimental data overwhelmingly shows that employing optimally sized fragments of 250-400 bp, delivered via advanced vectors like TRV-2b through high-efficiency methods such as vacuum infiltration, creates a synergistic effect that maximizes silencing efficacy. As functional genomics continues to probe the complexities of plant genomes, adhering to these optimized strategies for VIGS will ensure that researchers generate robust, reliable, and biologically meaningful data, effectively bridging the gap between gene sequence prediction and confirmed gene function.
Addressing Species-Specific Challenges in Woody Plants and Cereals
The validation of gene function is a cornerstone of modern plant molecular biology, enabling breakthroughs in crop improvement and basic science. Among the various techniques available, virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics tool for transiently knocking down gene expression without the need for stable transformation. However, its application across diverse plant species—particularly when comparing long-lived woody plants with annual cereals—presents distinct challenges and considerations. VIGS can be directly applied to infect target plants to avoid the complexity of plant genetic transformation and regeneration systems, and can infect plants with different genetic backgrounds simultaneously [48]. This advantage can largely shorten the period of gene function identification compared to stable transformation approaches like CRISPR/Cas9.
This guide provides a comparative analysis of VIGS implementation in woody and cereal species, synthesizing experimental data and optimized protocols to assist researchers in selecting appropriate methodologies for their specific plant systems. By addressing the species-specific biological and technical constraints, we aim to enhance the efficiency and reliability of gene function validation across the plant kingdom.
VIGS Mechanism and Workflow
VIGS operates by harnessing the plant's innate RNA-mediated defense mechanisms against viruses. When a viral vector carrying a fragment of a plant gene is introduced, the plant's silencing machinery processes this into small interfering RNAs (siRNAs) that target both the viral genome and corresponding endogenous mRNAs for degradation. This process involves key components including dicer-like enzymes, RNA-dependent RNA polymerases, and argonaute proteins, ultimately leading to sequence-specific degradation of target transcripts [62].
The tobacco rattle virus (TRV) has become one of the most widely used VIGS vectors due to its broad host range, effective systemic movement, and ability to target meristematic tissues [48]. The TRV genome comprises two positive-sense, single-stranded RNAs: RNA1 contains elements for viral replication and movement, while RNA2 can be engineered to carry host gene fragments without affecting infectivity in controlled conditions [48].
The following diagram illustrates the generalized experimental workflow for establishing a VIGS system in a new plant species, incorporating key optimization steps:
Comparative Analysis of VIGS Applications
Efficiency Metrics Across Species
VIGS efficiency varies considerably across plant species, influenced by factors including inoculation method, viral vector selection, and host-pathogen interactions. The table below summarizes key performance metrics documented in recent studies:
Table 1: Comparative VIGS Efficiency Across Plant Species
| Plant Species | Viral Vector | Optimal Infiltration Method | Target Gene | Silencing Efficiency | Silencing Duration | Key Optimization Factors |
|---|---|---|---|---|---|---|
| Walnut ('Qingxiang') | TRV | Apical meristem wounding | JrPDS | 48% | 14-21 days | Seedling age (2-3 true leaves), OD600=0.6, fragment length=255 bp [48] |
| Petunia ('Picobella Blue') | TRV | Mechanical wounding of shoot apical meristem | CHS, PDS | 69% (CHS), 28% (PDS) | 14-16 days | Temperature (20°C day/18°C night), cultivar selection [62] |
| Passion fruit | TRV | Vacuum infiltration (0.8 KPA, 10 min) | PePDS | 46.7% | 14-16 days | OD600=0.8, cotyledon stage inoculation [63] |
| Citrus | CLBV | Not specified | PDS | Varies by construct | Several months | Vector stability, secondary siRNA generation [64] |
| Grapevine (Vitis vinifera) | GVA | Agrobacterium-mediated via roots | PDS | Not quantified | Not specified | Micropropagated plantlets, root inoculation [65] |
Species-Specific Protocol Optimization
Successful implementation of VIGS requires careful optimization of multiple parameters specific to each plant system. The following experimental data highlight how these factors influence silencing efficiency:
Table 2: Species-Specific Optimization Parameters for VIGS
| Optimization Factor | Walnut | Petunia | Passion Fruit | Citrus |
|---|---|---|---|---|
| Plant Developmental Stage | 2-3 true leaves [48] | 3-4 weeks after sowing [62] | Cotyledon stage (~20 days) [63] | Successive flushes [64] |
| Agrobacterium Density (OD600) | 0.6 [48] | Not specified | 0.8 [63] | Not specified |
| Fragment Length | 255 bp optimal [48] | Not specified | 283 bp (PePDS) [63] | 58-nt hairpin effective [64] |
| Temperature Regime | 24°C [48] | 20°C day/18°C night [62] | 27°C [63] | Not specified |
| Optimal Cultivar | 'Qingxiang' > 'Xiangling' [48] | 'Picobella Blue' [62] | Not specified | Not specified |
Detailed Experimental Protocols
VIGS Implementation in Walnut
The establishment of an efficient TRV-mediated VIGS system in walnut (Juglans regia L.) represents a significant advancement for functional genomics in this recalcitrant species. The following protocol achieved up to 48% silencing efficiency:
Vector Construction:
- Identify target gene sequence from walnut genome (e.g., JrPDS, LOC108996675)
- Select 255 bp fragment from CDS region (designated JrPDS255) based on optimal fragment length testing [48]
- Clone into pTRV2 vector using appropriate restriction enzymes (EcoRI and XhoI)
- Transform into Agrobacterium tumefaciens strain GV3101
Plant Material Preparation:
- Soak seeds of walnut cultivars 'Qingxiang' and 'Xiangling' in water for 7 days
- Incubate in sand until germination occurs
- Transfer germinated seeds to seedling pots
- Maintain in light incubator at 24°C with 16h/8h light/dark cycle, light intensity 100 μmol m⁻² s⁻¹ [48]
Infiltration Methods:
- Apical meristem wounding: Most effective method, showing tiny light spots and leaf edge yellowing at 14 days post-infiltration (dpi)
- Seed soaking: Less efficient, showing minor photobleaching only at leaf edges
- Seedling spraying: Least efficient with minimal phenotypic changes [48]
Efficiency Assessment:
- Monitor photobleaching phenotype from 14 dpi
- Quantify expression levels via qRT-PCR with walnut actin gene as internal reference
- Calculate relative expression using 2(-Delta Delta C(T)) method [48]
VIGS Optimization in Petunia
Substantial improvements to VIGS efficiency in petunia were achieved through systematic optimization across six parameters:
Inoculation Technique Comparison:
- Mechanical wounding of shoot apical meristems induced most effective and consistent silencing
- Superior to leaf infiltration and other methods [62]
Cultivar Screening:
- Evaluation of ten cultivars revealed significant variation in silencing efficiency
- 'Picobella Blue' exhibited 1.8-fold higher CHS silencing efficiency in corollas compared to other cultivars [62]
Temperature Optimization:
- 20°C day/18°C night temperatures induced stronger gene silencing
- Outperformed 23°C/18°C or 26°C/18°C regimes [62]
Developmental Stage:
- More pronounced silencing in plants inoculated at 3-4 versus 5 weeks after sowing [62]
Vector Improvement:
- Developed pTRV2-sGFP control construct containing green fluorescent protein fragment
- Eliminated severe necrosis and stunting observed with empty pTRV2 vector
- Enabled proper phenotyping of silenced plants [62]
Woody Plant vs. Cereal Considerations
The biological and technical challenges differ substantially between woody plants and cereals when implementing VIGS:
Woody Plant Specific Challenges
Woody species like walnut, grapevine, and fruit trees present unique obstacles for VIGS implementation. These plants often lack robust tissue culture and genetic transformation systems, making transient approaches like VIGS particularly valuable [48]. Many studies related to walnut abiotic stress resistance have focused on biomass, morphological, physiological, and biochemical differences, whereas molecular mechanisms are lacking due to these technical limitations [48].
Long generation times and complex architecture necessitate VIGS vectors with sustained activity. The Citrus leaf blotch virus (CLBV)-based vectors offer advantage in this regard, as they remain stable and induce VIGS in successive flushes for several months [64]. This stability provides an important genomic tool for analyzing gene function by reverse genetics in long-lived plants.
Meristematic targeting is another crucial factor. TRV vectors are prominent in exploring gene functions in forest trees species due to their capability to suppress the activity of genes expressed in plant meristems [48]. This enables functional studies of genes involved in development and perennial growth patterns.
Cereal Adaptation Considerations
While the search results provided limited specific data on cereal VIGS applications, general considerations can be noted based on comparative analysis. Cereal species often exhibit different efficiency in generating secondary siRNAs, which significantly impacts VIGS effectiveness. For instance, CLBV-based vectors induced gene silencing in citrus but not in Nicotiana benthamiana, potentially due to differences in the generation of secondary siRNAs in both species [64].
Cereal-specific viral vectors may be required for optimal performance, as different viruses have varying host ranges and movement capabilities. To date, multiple viruses have been modified into VIGS vectors, including cucumber mosaic virus (CMV), tobacco mosaic virus (TMV), tomato leaf curl virus (ToLCV), and tobacco rattle virus (TRV) [48].
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of VIGS requires carefully selected reagents and vectors tailored to specific plant systems. The following table summarizes key solutions for establishing VIGS in challenging species:
Table 3: Essential Research Reagents for VIGS Implementation
| Reagent/Vector | Specifications | Function & Application Notes |
|---|---|---|
| TRV Vectors | pTRV1 (pYL192) and pTRV2 (pYL156) [62] | Most widely used VIGS system; effective across diverse species including woody plants |
| Alternative Vectors | GVA (Grapevine virus A) [65], CLBV (Citrus leaf blotch virus) [64] | Species-specific vectors for plants where TRV is ineffective |
| Agrobacterium Strain | GV3101 [48] [63] | Standard strain for plant transformation; optimized for VIGS delivery |
| Marker Genes | PDS (Phytoene desaturase) [48], CHS (Chalcone synthase) [62] | Visual indicators of silencing efficiency through photobleaching or pigment loss |
| Infiltration Buffer | 10 mM MES, 200 μM AS, 10 mM MgCl₂, pH 5.6 [63] | Optimized for Agrobacterium-mediated delivery of viral constructs |
| Control Constructs | pTRV2-sGFP (contains GFP fragment) [62] | Prevents viral symptoms from empty vector; proper experimental control |
Technical Challenges and Troubleshooting
Even with optimized protocols, researchers may encounter several technical challenges when implementing VIGS:
Variable Silencing Efficiency: Efficiency can vary considerably across cultivars, as demonstrated in walnut where 'Qingxiang' showed better performance than 'Xiangling' [48]. To address this, screen multiple cultivars and optimize parameters for each specific genotype.
Viral Symptoms: Severe necrosis and stunting can occur with empty vectors, particularly in petunia [62]. The solution is to use control vectors containing non-plant gene inserts (e.g., GFP) rather than empty vectors.
Species-Specific Vector Compatibility: Vectors that work in one species may fail in others due to differences in siRNA processing [64]. When working with new species, test multiple vector systems and consider designing species-specific vectors.
Transient Nature: Silencing is typically transient, lasting 2-3 weeks in many systems [63]. For longer-term studies, consider repeated inoculation or alternative vector systems like CLBV that provide sustained silencing [64].
VIGS technology has revolutionized functional genomics in species recalcitrant to stable transformation, particularly woody plants. The experimental data and protocols presented here demonstrate that success depends on systematic optimization of multiple species-specific parameters including plant developmental stage, inoculation method, viral vector selection, and environmental conditions. While TRV-based vectors show remarkable versatility across diverse species, alternative vectors like GVA and CLBV offer solutions for particularly challenging systems.
The comparative analysis reveals that woody plants generally require more intensive optimization but can achieve silencing efficiencies comparable to model species when protocols are properly adapted. As VIGS methodologies continue to evolve, they will increasingly enable rapid gene function validation across the plant kingdom, accelerating both basic research and crop improvement efforts.
Understanding the function of plant genes is a central task in molecular biology and is foundational for modern breeding and genetic engineering. Virus-Induced Gene Silencing (VIGS) has emerged as a powerful functional genomics tool that enables transient, sequence-specific gene knockdown, allowing researchers to link genes to functions by observing resulting phenotypic changes [3]. A critical advantage of VIGS is its applicability to plant species recalcitrant to stable genetic transformation, such as pepper, watermelon, and luffa, where it often serves as the primary tool for high-throughput functional screening [3] [50] [4].
The efficacy of VIGS, as well as the phenotypic outcomes of gene function studies, are profoundly influenced by environmental conditions. Temperature, humidity, and photoperiod constitute key environmental signals that plants process simultaneously, with specific combinations often providing more developmental information than any single factor alone [66]. This guide objectively compares the effects of these environmental controls on plant development and gene expression, providing experimental data and methodologies relevant to designing robust VIGS and functional genomics experiments. A comprehensive understanding of these parameters is essential for distinguishing genuine genetic effects from environmentally induced phenotypic variation, thereby enhancing the validity of functional gene characterization.
Comparative Analysis of Environmental Factors
The following sections provide a detailed, data-driven comparison of how temperature, humidity, and photoperiod influence plant growth, development, and metabolic processes, with direct implications for the design and interpretation of gene function studies.
Temperature Effects on Development and Metabolism
Temperature significantly influences a wide range of physiological processes, from fundamental metabolism to developmental transitions such as flowering. The effects are often genotype-specific and can interact strongly with other environmental cues.
Table 1: Temperature Effects on Plant Development and Metabolism
| Plant Species | Temperature Conditions | Key Physiological Effects | Impact on Key Traits |
|---|---|---|---|
| Cannabis sativa (low-THC hemp 'V4') [67] | Moderate (27°C day/21°C night) vs. Constant (24°C) | Optimized cannabinoid synthesis | ↑ CBD-A (28.75%), ↑ CBC-A (43.6%), ↑ THC-A (41.3%) |
| Cannabis sativa (THC/CBD-rich) [67] | High (31°C/27°C) vs. Lower (25°C/21°C) | Disrupted inflorescence development, reduced biomass | ↓ Total cannabinoid yield from >400/200 g m⁻² to <100 g m⁻² |
| Arabidopsis thaliana [66] | Low (15°C) vs. Higher (20°C/25°C) | Modulated flowering time via PHOT2/CAMTA2 pathway | ↑ Leaf number at flowering in phot2 mutants at 15°C |
| Rice (Oryza sativa ssp. japonica) [68] | Low (22°C) vs. High (28°C) during PSP | Altered heading date; interaction with photoperiod | Thermosensitivity increased threefold under short-day conditions |
The genetic mechanisms underlying temperature sensitivity are becoming clearer. Research in Arabidopsis has revealed that the blue light photoreceptor PHOTOTROPIN 2 (PHOT2) integrates with low-temperature information to control flowering time. Mutants lacking PHOT2 flower later than controls specifically in low ambient temperature (15°C), a phenotype that requires blue light and is blocked by removal of its signaling partner, NPH3 [66]. Furthermore, a non-additive genetic interaction between PHOT2 and the transcription factor CAMTA2 suggests this module acts as a light-temperature coincidence detector, buffering development against environmental fluctuations [66].
In rice, the interaction between temperature and photoperiod is critical for the heading date. Integrative transcriptomic and metabolomic analyses have shown that thermosensitivity increases threefold under short-day conditions, while photosensitivity is enhanced under high temperature [68]. This interaction underscores the importance of controlling both variables precisely in functional studies.
Photoperiod and Light Quality Interactions
Photoperiod governs developmental transitions by providing predictable seasonal information, but its effects are often modulated by light quality and temperature.
Table 2: Photoperiod and Light Quality Effects on Plant Development
| Plant Species | Light Regime | Key Physiological Effects | Impact on Key Traits |
|---|---|---|---|
| Cannabis sativa [67] | 12-hour photoperiod | Optimized floral initiation and CBD production | ↑ CBD concentration and dry biomass yield |
| Rice (Oryza sativa) [68] | Short (12h) vs. Long (14.5h) days | Altered photosensitivity; interaction with temperature | Photosensitivity enhanced under high temperature |
| Cannabis sativa (cuttings) [69] | 20/4 h (light/dark) with blue light | Promoted adventitious root formation | Enhanced rooting in stem cuttings |
The molecular basis of photoperiodic control is well-established in rice, where short-day conditions activate florigen genes like Hd3a and RFT1 to induce panicle initiation [68]. However, the integration of photoperiod with temperature sensing involves complex regulatory networks. For example, in Arabidopsis, PHOT2 signaling combines with low-temperature input through the transcription factor CAMTA2, creating a system that fine-tunes flowering based on concurrent light and temperature conditions [66].
Relative Humidity Impacts on Plant Physiology
Relative humidity (RH) primarily affects plant water balance and transpiration, with significant consequences for growth, development, and specialized metabolism.
Table 3: Effects of Relative Humidity on Plant Growth and Metabolism
| Plant Species | RH Conditions | Key Physiological Effects | Impact on Key Traits |
|---|---|---|---|
| Cannabis sativa [67] | High RH (78-98%) vs. Low RH (37-58%) | Reduced vapor pressure deficit (VPD), delayed development | ↓ Total biomass (-75.3%), ↓ flower biomass (-71.0%), delayed flowering (3 weeks) |
| Cannabis sativa [67] | High RH (78-98%) | Suppressed cannabinoid biosynthesis | ↓ CBD-A (~4.9-fold), ↓ CBD (~3.2-fold), ↓ CBC-A (~13-fold) |
| Cannabis sativa (cuttings) [69] | High RH (90%) | Promoted adventitious rooting | Enhanced root formation in stem cuttings |
Cultivation under high RH (78-98%) significantly reduces the vapor pressure deficit (VPD), creating suboptimal conditions for cannabis. This environment leads to substantial reductions in biomass, delayed flowering, and severely suppressed cannabinoid accumulation [67]. Conversely, for propagation, high RH (90%) promotes adventitious root formation in stem cuttings by reducing transpirational water loss and facilitating auxin transport [69]. This demonstrates that optimal RH levels are often trait-specific and depend on the developmental stage.
Experimental Protocols for Environmental Studies
High-Throughput VIGS Protocol for Abiotic Stress Studies
A robust VIGS protocol has been developed for high-throughput functional characterization of genes involved in abiotic stress tolerance [70]. This methodology employs VIGS-based gene silencing in leaf disks combined with simple stress imposition techniques.
Key Methodology Steps:
- Gene Silencing: Clone 300-580 bp fragments of target genes into the pTRV2 VIGS vector and silence them in Nicotiana benthamiana via standard agroinfiltration [70].
- Leaf Disk Excision: At 20 days post-inoculation (dpi), punch leaf disks from newly developed silenced leaves.
- Stress Application: Expose leaf disks to abiotic stresses by incubating them on MS medium or callus induction medium supplemented with stress-inducing agents.
- Phenotyping: Quantify stress effects by measuring chlorophyll fluorescence, ion leakage, or biomass accumulation.
This system allows for efficient screening of genes involved in multi-stress tolerance, including salinity, oxidative stress, and temperature extremes [70]. The protocol is particularly valuable because gene silencing continues in excised leaf disks for more than six weeks, enabling extended phenotyping without maintaining whole plants [70].
Controlled Environment Protocol for Photoperiod-Temperature Interactions
To dissect the interaction between photoperiod and temperature on rice heading date, a controlled-environment study was conducted with precise environmental control [68].
Key Methodology Steps:
- Plant Materials: Use contrasting cultivars (e.g., early-maturing 'Odae' and mid-late-maturing 'Saenuri').
- Treatment Application: Apply combinations of temperature (22°C vs. 28°C) and photoperiod (12h vs. 14.5h) during the photosensitive period (PSP).
- Molecular Analysis: Conduct integrative transcriptomic and metabolomic analyses to identify key regulators.
- Metabolite Application: Validate findings through exogenous application of identified metabolites (e.g., 250-500 μM glycerate).
This approach revealed that glycerate, a pivotal intermediate in photorespiration and glycolysis, showed an inverse correlation with days to heading and accumulated more strongly under short-day and high-temperature conditions [68]. Exogenous glycerate application accelerated heading by 4-5 days, confirming its functional role in floral induction [68].
Signaling Pathways and Genetic Networks
Environmental signals are processed through complex genetic networks that integrate multiple cues to coordinate development. The following diagrams illustrate key pathways mediating these responses.
PHOT2-CAMTA2 Blue Light-Temperature Coincidence Detection
Figure 1: Genetic architecture of the PHOT2-CAMTA2 blue light-temperature coincidence detector. This module integrates blue light intensity information from PHOT2 with low-temperature signals from CAMTA2 to fine-tune flowering time in Arabidopsis. CAMTA2 promotes expression of EHB1, which interacts with NPH3, potentially serving as the point of temperature input into the PHOT-NPH3 light signaling pathway [66].
VIGS Experimental Workflow for Environmental Studies
Figure 2: VIGS experimental workflow for environmental studies. This workflow illustrates the key steps in using VIGS to study gene function under controlled environmental conditions. Critical parameters include temperature, photoperiod, and humidity, which must be strictly controlled during plant growth and stress application to ensure reproducible results [3] [50] [70].
Research Reagent Solutions
The following table details essential research reagents and methodologies used in environmental control studies and VIGS experiments.
Table 4: Key Research Reagents and Methodologies for Environmental Plant Studies
| Reagent/Methodology | Specifications | Research Application | Key References |
|---|---|---|---|
| TRV-VIGS Vector | Bipartite system (TRV1/TRV2); broad host range | Functional gene analysis in Solanaceae (pepper, tomato) | [3] |
| CFMMV-VIGS Vector | pCF93 vector; 93 bp upstream of CP; monopartite | High-throughput gene validation in cucurbits (watermelon) | [50] |
| CGMMV-VIGS Vector | pV190 vector; BamHI insertion site | Gene function studies in cucurbits (luffa, cucumber) | [4] |
| Leaf-Disk VIGS Assay | Excised tissues on MS/CIM medium | High-throughput abiotic stress tolerance screening | [70] |
| Controlled Environment Chambers | Precise control of light, temperature, humidity | Studying photoperiod-temperature interactions (rice, cannabis) | [68] [69] |
| HPLC Cannabinoid Profiling | Quantification of 14+ cannabinoids | Assessment of environmental effects on specialized metabolism | [67] |
| Integrative Transcriptomic/Metabolomic Analysis | RNA-seq combined with metabolite profiling | Identification of regulatory networks in heading date (rice) | [68] |
Environmental controls—temperature, humidity, and photoperiod—profoundly influence plant development, specialized metabolism, and the outcomes of functional genomics studies. Temperature and photoperiod frequently interact in complex ways, as demonstrated by the PHOT2-CAMTA2 coincidence detection module in Arabidopsis and the glycerate-mediated heading regulation in rice [66] [68]. Humidity significantly affects both vegetative growth and metabolic profiles, with high RH reducing cannabinoid yields in cannabis while promoting adventitious rooting in cuttings [67] [69].
For researchers employing VIGS and knockout studies, strict control of these environmental parameters is essential for distinguishing genuine gene function from environmentally induced variation. The experimental protocols and reagents detailed in this guide provide a foundation for designing robust functional genomics experiments that account for these critical environmental interactions. Future research integrating multi-omics approaches with controlled environment studies will further illuminate the complex networks through which plants integrate environmental signals to optimize growth and development.
Employing Viral Suppressors of RNA Silencing (VSRs) to Boost VIGS Efficiency
Virus-induced gene silencing (VIGS) has emerged as a powerful functional genomics tool that allows for rapid characterization of gene function by leveraging the plant's innate RNA silencing machinery. However, the efficacy of VIGS is inherently limited by the plant's defense mechanisms that recognize and degrade viral RNA. Viral Suppressors of RNA Silencing (VSRs) are specialized proteins encoded by plant viruses to counteract these host defenses, thereby enabling viral accumulation and spread. The strategic deployment of these VSRs within VIGS protocols presents a significant opportunity to enhance silencing efficiency, particularly in plant species that are traditionally recalcitrant to established VIGS methods.
The biological foundation of VIGS is the plant's post-transcriptional gene silencing (PTGS) mechanism, an antiviral defense system. This process involves Dicer-like enzymes (DCL) cleaving viral double-stranded RNA replication intermediates into 21- to 24-nucleotide small interfering RNAs (siRNAs). These siRNAs are then incorporated into the RNA-induced silencing complex (RISC), which guides sequence-specific degradation of complementary viral mRNA [3]. VSRs interfere with this pathway at various points, and their exploitation is particularly valuable for functional studies in species like pepper (Capsicum annuum L.), where stable genetic transformation remains challenging and VIGS often serves as the primary tool for high-throughput functional screening [3].
Key VSRs and Their Mechanisms of Action
Major Viral Suppressors and Functional Characteristics
Different VSRs have been characterized and utilized to enhance VIGS across various plant-virus systems. Research has demonstrated that the efficacy of these suppressors varies among plant species, a consideration exploited to enhance VIGS efficiency [3].
Table 1: Key Viral Suppressors of RNA Silencing (VSRs) and Their Properties
| VSR Name | Viral Origin | Mechanism of Action | Reported Impact on VIGS Efficiency |
|---|---|---|---|
| P19 | Tomato bushy stunt virus (TBSV) | Binds and sequesters siRNA duplexes, preventing RISC assembly [3]. | One of the most potent enhancers; widely used to boost silencing in multiple VIGS systems. |
| HC-Pro | Potato virus Y (PVY) | Inhibits siRNA amplification and may interfere with Dicer-like activity [71] [3]. | Enhances silencing persistence and systemic spread. |
| C2b | Cucumber mosaic virus (CMV) | Suppresses silencing initiation and may interfere with systemic signaling [3]. | Used to augment VIGS constructs, especially in non-model hosts. |
| AC2 | Geminiviruses | Functions as a transcriptional activator and suppresses the silencing pathway [71]. | Can enhance VIGS efficiency in systems using geminivirus-based vectors. |
| P15 | Peanut clump virus | Not fully elucidated, but involved in suppression of systemic silencing. | Contributes to viral movement and can aid in systemic VIGS. |
| P38 | Turnip crinkle virus | Binds and inhibits Dicer-like proteins (e.g., DCL4) [71]. | Effective in increasing the accumulation of viral vectors. |
Molecular Pathways of VSR Activity
The following diagram illustrates the plant RNA silencing pathway and the points where key VSRs, such as P19 and HC-Pro, exert their suppressive effects to enhance VIGS.
Diagram 1: Plant RNA Silencing Pathway and VSR Intervention Points. The green nodes represent the natural host silencing pathway, while the red nodes indicate the points where specific VSRs inhibit the process to enhance VIGS efficiency.
Experimental Validation and Performance Data
Quantitative Assessment of VSR-Enhanced VIGS
The integration of VSRs into VIGS protocols has been quantitatively shown to improve key performance metrics. The enhancements are most evident in the strength and duration of the silencing phenotype, the speed of onset, and the consistency of results across a population of treated plants. For instance, in pepper, the use of VSRs like P19 has been critical for achieving reliable silencing of genes involved in complex traits such as fruit quality, pungency, and resistance to biotic and abiotic stresses [3].
Table 2: Comparative Performance of VIGS with and without VSR Enhancement
| Performance Metric | Standard VIGS | VIGS + VSR (e.g., P19) | Experimental Context |
|---|---|---|---|
| Silencing Onset Time | 2-3 weeks | 1-2 weeks | TRV-based system in Nicotiana benthamiana [3]. |
| Phenotype Strength | Variable, often moderate | Strong, highly penetrant | Silencing of PDS leading to photobleaching. |
| Silencing Duration | 2-4 weeks | 4-6 weeks, sometimes longer | In meristematic tissues, which are often difficult to silence. |
| Systemic Spread | Limited in some species | Enhanced, more uniform | Silencing in newly emerged leaves. |
| Experiment-to-Experiment Consistency | Can be variable | High reproducibility | Functional screening of resistance genes in pepper [3]. |
Case Study: Validating Gene Function in Cotton NBS Genes
A compelling application of optimized VIGS was demonstrated in the functional validation of a nucleotide-binding site (NBS) domain gene, a key resistance gene class. Researchers investigated the role of these genes in tolerance to cotton leaf curl disease (CLCuD). Expression profiling identified specific orthogroups (OGs), including OG2, that were upregulated in tolerant cotton accessions under biotic stress [72].
To functionally validate the role of a gene from OG2 (GaNBS), researchers performed VIGS in resistant cotton. The critical experimental detail was the use of a VIGS vector based on the Cotton leaf crumple virus (CLCrV), a geminivirus [3]. The protocol involved:
- Vector Construction: Cloning a fragment of the target GaNBS gene into the CLCrV VIGS vector.
- Agroinfiltration: Introducing the recombinant vector into Agrobacterium tumefaciens and infiltrating the bacterial culture into cotyledons of resistant cotton plants.
- Phenotypic Assessment: After virus spread, plants were challenged with the cotton leaf curl disease virus.
- Result: The GaNBS-silenced plants, which would have exhibited an enhanced VIGS response if supplemented with a VSR like AC2, showed a significant increase in viral titer compared to controls. This result confirmed the critical role of GaNBS in the plant's defense response, a finding that was only possible with an efficient VIGS system [72].
Essential Research Reagent Solutions
The successful implementation of VSR-enhanced VIGS relies on a toolkit of specialized reagents and biological materials.
Table 3: Essential Research Reagent Solutions for VSR-Enhanced VIGS
| Reagent / Material | Function / Purpose | Examples / Notes |
|---|---|---|
| Viral Vectors with VSR | Backbone for delivering the target gene fragment and the VSR. | TRV, BBWV2, CLCrV, ACMV; often used with co-expressed P19 or HC-Pro [3]. |
| Agrobacterium Strains | Delivery vehicle for the VIGS construct into plant cells. | GV3101 is commonly used for agroinfiltration [21]. |
| VSR Expression Plasmids | Source of the silencing suppressor gene. | Plasmids expressing P19, HC-Pro, or C2b can be co-infiltrated or integrated into the VIGS vector [3]. |
| Plant Growth Formulations | To maintain robust plant health post-infiltration, a stressor. | Controlled-environment chambers with optimized temperature, humidity, and photoperiod are critical [3]. |
| Validation Tools | To confirm and quantify the extent of gene silencing. | qRT-PCR primers for the target gene; antibodies for phenotypic or protein-level analysis. |
Integrated Experimental Workflow for VSR-Enhanced VIGS
The following diagram outlines a standardized protocol for implementing a VSR-boosted VIGS experiment, from design to functional validation.
Diagram 2: Integrated Workflow for a VSR-Boosted VIGS Experiment. The blue nodes represent the core experimental steps, while the yellow notes list critical parameters for success at each stage.
The strategic use of Viral Suppressors of RNA Silencing represents a sophisticated and highly effective method for augmenting the efficiency of Virus-Induced Gene Silencing. By neutralizing components of the host plant's defensive RNAi machinery, VSRs like P19 and HC-Pro directly address the primary limitation of VIGS, enabling stronger, faster, more persistent, and more reliable silencing. As functional genomics continues to advance in non-model crops and species resistant to transformation, the integration of VSRs into standard VIGS protocols will be indispensable for high-throughput gene validation, pathway elucidation, and ultimately, for accelerating crop improvement programs.
From Phenotype to Function: Validation and Technology Integration
Molecular validation is a cornerstone of modern plant functional genomics, enabling researchers to link genetic sequences to biological functions. As genome sequencing technologies advance, generating vast amounts of genomic data, the need for efficient and reliable methods to validate gene function has become increasingly important. This guide focuses on two pivotal aspects of gene validation: molecular verification using Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) and phenotypic assessment through Virus-Induced Gene Silencing (VIGS) and related technologies. RT-qPCR provides precise quantification of gene expression changes, while VIGS enables rapid functional characterization through transient gene silencing. The integration of these methods offers a powerful framework for comprehensive gene function analysis, particularly in plant species where stable transformation remains challenging. This article objectively compares current protocols and their performance parameters, providing experimental data and methodological details to guide researchers in selecting appropriate validation strategies for their specific applications.
RT-qPCR in Gene Expression Validation
Principles and Workflow
Reverse Transcription Quantitative PCR (RT-qPCR) serves as a fundamental tool for validating gene expression changes in functional genomics studies. This technique enables precise quantification of transcript abundance by first converting RNA to complementary DNA (cDNA) via reverse transcription, followed by quantitative PCR amplification with sequence-specific primers. The reliability of RT-qPCR data heavily depends on proper experimental design, including appropriate controls and normalization strategies [73].
A critical advancement in RT-qPCR validation has been the recognition that housekeeping genes, traditionally used as internal controls, can exhibit significant expression variability under different experimental conditions. Recent studies have emphasized the necessity of experimentally validating reference genes for each specific biological system. For instance, in adipocyte research, HPRT and HMBS have been identified as more stable reference genes compared to the traditionally used GAPDH and Actb, which showed significant variability [73]. Similarly, in aging studies using African turquoise killifish, cyc1, oaz1a, and gusb demonstrated greater stability than gapdh and actb [74]. These findings highlight the importance of preliminary validation rather than presuming the stability of commonly used reference genes.
Experimental Protocol for Plant Gene Expression Analysis
The following protocol outlines the key steps for conducting RT-qPCR analysis in plant gene function studies:
RNA Extraction: Harvest plant tissue (50-100 mg) and immediately freeze in liquid nitrogen. Extract total RNA using commercial kits (e.g., RNeasy Plus Mini Kit, HiPure Gel Pure DNA Mini Kit, or Eastep Super Total RNA Extraction Kit) following manufacturer instructions [75] [73] [4]. Determine RNA purity and concentration spectrophotometrically (A260/A280 ratio of ~2.0 is ideal) [73].
cDNA Synthesis: Use 1000 ng of total RNA for reverse transcription with RT master mix. Perform the reaction on a thermal cycler using manufacturer-recommended conditions [75].
qPCR Reaction Setup: Prepare 10 μL reactions containing: 2 μL cDNA, 0.2 μL forward primer (200 nM final concentration), 0.2 μL reverse primer (200 nM final concentration), 2.6 μL nuclease-free water, and 5 μL qPCR Master Mix [75].
Amplification Parameters: Program the thermal cycler as follows: initial denaturation at 95°C for 3 minutes; 40 cycles of 95°C for 5 seconds and 61°C for 30 seconds; followed by melting curve analysis from 65°C to 95°C [75].
Data Analysis: Calculate fold-change differences using the 2^(-ΔΔCq) method. Validate primer efficiencies (95-101% ideal) using standard curves from serial cDNA dilutions. Include three biological replicates with technical triplicates for statistical robustness [75].
Table 1: Comparison of RT-qPCR Experimental Parameters Across Studies
| Application Context | Reference Genes Validated | Most Stable Reference Genes | Key Normalization Recommendations |
|---|---|---|---|
| 3T3-L1 Adipocytes [73] | GAPDH, Actb, HPRT, HMBS, 18S, 36B4 | HPRT, HMBS, 36B4 | Use HPRT and HMBS as a stable pair; include 36B4 for triplet normalization |
| Killifish Brain Aging [74] | actb, cyc1, gapdh, gusb, oaz1a, tbp | cyc1, oaz1a, gusb | Use at least two of the top-ranked RGs with sex-specific consideration |
| Macrophage Polarization [75] | 18S rRNA | 18S rRNA | Single reference gene used after validation |
Phenotypic Assessment in Gene Function Studies
Virus-Induced Gene Silencing (VIGS) Principles
Virus-Induced Gene Silencing (VIGS) is a powerful reverse genetics tool that leverages the plant's natural RNA silencing machinery to investigate gene function. The technology utilizes recombinant viral vectors carrying fragments of plant genes to trigger sequence-specific mRNA degradation through post-transcriptional gene silencing (PTGS) [4] [3]. When plants are infected with these engineered viruses, the plant's defense mechanism recognizes and processes the viral double-stranded RNA replication intermediates into small interfering RNAs (siRNAs). These siRNAs are then incorporated into the RNA-induced silencing complex (RISC), which guides the cleavage of complementary endogenous mRNA transcripts, resulting in knock-down of the target gene and potentially revealing phenotypic consequences [3].
The fundamental advantage of VIGS lies in its ability to rapidly generate loss-of-function phenotypes without the need for stable transformation, which remains challenging in many plant species. Since its initial demonstration in 1995 using a Tobacco mosaic virus vector carrying a phytoene desaturase (PDS) gene fragment, VIGS has been adapted for numerous plant species using various viral vectors [3]. The visual silencing of marker genes like PDS (causing photobleaching) and chalcone synthase (CHS, causing loss of pigmentation) provides straightforward indicators of silencing efficiency [62] [4].
VIGS Experimental Protocol
The following protocol details the establishment of a VIGS system using a CGMMV-based vector in Luffa acutangula, with principles applicable to other plant-virus systems:
Vector Construction:
- Clone 300-500 bp fragment of target gene (e.g., PDS, TEN) using gene-specific primers with appropriate restriction sites [4].
- Insert fragment into multiple cloning site of viral vector (e.g., pV190 for CGMMV system) using restriction enzymes (BamHI) or homologous recombination [4].
- Transform construct into Agrobacterium tumefaciens strain (e.g., GV3101) for delivery [4].
Plant Material Preparation:
Agroinfiltration:
- Culture Agrobacterium carrying VIGS construct in YEP medium with appropriate antibiotics (50 mg/L Kan, 25 mg/L Rif) to OD600 = 0.6-0.8 [4].
- Pellet bacteria by centrifugation and resuspend in infiltration buffer (10 mM MgCl₂, 10 mM MES, 200 μM AS) to OD600 = 0.8-1.0 [4].
- Incubate suspension at room temperature for 2+ hours [4].
- Inoculate by making small holes in cotyledons or true leaves with syringe needle and infiltrating agrobacterium suspension from abaxial leaf side using needleless syringe [4].
Post-Inoculation Care:
Silencing Efficiency Assessment:
VIGS Vector Systems Comparison
Various viral vectors have been developed for VIGS applications, each with distinct advantages and limitations. The selection of an appropriate vector system depends on the host plant species, target tissue, and required duration of silencing.
Table 2: Comparison of VIGS Vector Systems and Applications
| Vector System | Virus Type | Host Range | Key Features | Silencing Duration | Example Applications |
|---|---|---|---|---|---|
| Tobacco Rattle Virus (TRV) [62] [3] | RNA virus | Broad (Solanaceae, Arabidopsis, etc.) | Efficient systemic movement; targets meristematic tissues | 3-8 weeks | Petunia (CHS, PDS), Tomato, Pepper |
| Cucumber Green Mottle Mosaic Virus (CGMMV) [4] | RNA virus | Cucurbits (cucumber, luffa, bottle gourd) | Minimal viral symptoms; effective in cucurbits | 3-6 weeks | Luffa (PDS, TEN), Watermelon, Cucumber |
| Apple Latent Spherical Virus (ALSV) [4] | RNA virus | Broad (6 cucurbit species+) | Mild or no symptoms; broad host range | 4-8 weeks | Various cucurbit species |
| Tobacco Ringspot Virus (TRSV) [4] | RNA virus | Melon, watermelon, cucumber, N. benthamiana | - | - | Melon, watermelon, cucumber |
| Bean Pod Mottle Virus (BPMV) [3] | RNA virus | Soybean | - | - | Soybean functional genomics |
| Cotton Leaf Crumple Virus (CLCrV) [3] | DNA virus | Cotton | - | - | Cotton gene function studies |
Integrated Validation Approaches
Combining Molecular and Phenotypic Assessment
Comprehensive gene validation requires integrating multiple approaches to establish robust genotype-phenotype relationships. The most convincing functional validation comes from correlating molecular changes (measured by RT-qPCR) with observable phenotypic effects (assessed through VIGS). Studies in transgenic rice lines demonstrate this integrated approach, where target gene insertion was confirmed through flanking sequence analysis, CRY1C and CRY2A gene expression was quantified using qRT-PCR, and phenotypic performance was evaluated through agronomic trait assessment [76].
Similar integrated approaches are used in non-plant systems, such as macrophage polarization studies, where RT-qPCR analysis of cytokine expression (IL-1β, IL-6, IL-10) is combined with flow cytometry for surface marker profiling (CD86, CD64, CD206) and fluorescence imaging using Di-4-ANEPPDHQ to characterize membrane properties [75]. This multi-faceted validation provides complementary evidence for functional changes.
Optimization Parameters for Validation Studies
Several critical factors influence the success and reliability of molecular validation studies:
Reference Gene Stability: As highlighted in multiple studies, experimental validation of reference genes is essential for accurate RT-qPCR normalization. Statistical algorithms (geNorm, NormFinder, BestKeeper, RefFinder) should be employed to identify the most stable internal controls under specific experimental conditions [73].
VIGS Efficiency Factors: Silencing efficiency depends on multiple parameters including insert size (300-500 bp optimal), agroinfiltration methodology, plant developmental stage, agroinoculum concentration (OD600 = 0.8-1.0), plant genotype, and environmental conditions (temperature, humidity, photoperiod) [62] [3].
Experimental Controls: Appropriate controls are crucial for both RT-qPCR (including no-template controls, reverse transcription controls) and VIGS (empty vector controls, non-inoculated plants, and marker gene silencing like PDS or CHS) [62] [4].
Temporal Considerations: The timing of analysis is critical—VIGS phenotypes typically appear 2-4 weeks post-infiltration, while gene expression analysis should be conducted when silencing is most pronounced [4].
Research Reagent Solutions
The following table outlines essential research reagents and materials commonly used in molecular validation experiments:
Table 3: Key Research Reagents for Molecular Validation Protocols
| Reagent / Material | Specific Example Products | Application Function | Experimental Considerations |
|---|---|---|---|
| Viral Vectors | pTRV1/pTRV2 (TRV), pV190 (CGMMV), pCF93 (CFMMV) [62] [4] [3] | Deliver target gene fragments to trigger RNA silencing | Select based on host compatibility; TRV has broad host range |
| Agrobacterium Strains | GV3101, K599 [21] [4] | Deliver viral vectors into plant tissues | Optimize OD600 (0.8-1.0); use with appropriate antibiotics |
| RNA Extraction Kits | RNeasy Plus Mini Kit, Eastep Super Total RNA Extraction Kit [75] [73] [4] | Isolate high-quality RNA for RT-qPCR | Assess RNA purity (A260/280 ~2.0); include DNase treatment |
| Reverse Transcription Kits | RT master mix (Takara) [75] | Convert RNA to cDNA for qPCR analysis | Use consistent input RNA (1000 ng recommended) |
| qPCR Master Mixes | qPCR Master Mix (Promega) [75] | Enable quantitative PCR amplification | Validate primer efficiencies (95-101% ideal) |
| Reference Genes | HPRT, HMBS, 36B4, cyc1, oaz1a, gusb [73] [74] | Normalize RT-qPCR data | Must validate stability for specific experimental conditions |
| Visual Marker Constructs | PDS (photobleaching), CHS (loss of pigmentation) [62] [4] | Visual indicators of silencing efficiency | Useful for optimizing VIGS protocols |
Molecular validation through RT-qPCR and phenotypic assessment represents a critical pathway for establishing gene function in plant genomics research. This comparison of current methodologies demonstrates that while RT-qPCR provides precise quantification of gene expression changes, its reliability depends heavily on proper experimental design, particularly the validation of stable reference genes. Meanwhile, VIGS technology offers a powerful approach for rapid functional characterization, especially in species where stable transformation remains challenging. The integration of these methods, along with emerging technologies such as virus-induced genome editing (VIGE) and nanoparticle-mediated transformation, continues to expand the toolbox available for plant functional genomics. As these technologies evolve, adherence to standardized protocols and implementation of appropriate controls will remain essential for generating robust, reproducible validation data. By understanding the comparative advantages, limitations, and optimal applications of each approach, researchers can select the most appropriate strategies for their specific gene validation needs.
VIGS as a Rapid Screening Tool Prior to Stable Transformation
In the field of plant functional genomics, stable genetic transformation has long been the cornerstone technology for validating gene function. This approach, which includes generating transgenic overexpression or RNA interference (RNAi) lines, provides heritable and stable phenotypic data. However, stable transformation is notoriously time-consuming and labor-intensive, particularly in agronomically important crops where transformation systems may be inefficient or genotype-dependent. For instance, soybean genetic transformation is characterized as "very time-consuming and laborious work" [77], while functional genomics in watermelon is challenging because it is "recalcitrant to genetic transformation" [50]. Similarly, tea plant research faces a "bottleneck" due to difficulties in establishing stable genetic transformation systems [78].
Virus-Induced Gene Silencing (VIGS) has emerged as a powerful alternative that bypasses many limitations of stable transformation. VIGS is a reverse genetics technology that leverages the plant's innate RNA-based antiviral defense mechanism to achieve transient silencing of target genes. When recombinant viral vectors carrying host gene fragments infect plants, the resulting sequence-specific RNA degradation silences corresponding endogenous mRNAs, enabling rapid functional analysis [79] [4]. This approach provides a rapid, high-throughput screening tool that allows researchers to prioritize the most promising candidate genes before committing to lengthy stable transformation programs.
How VIGS Works: Molecular Mechanisms and Technology
The Molecular Machinery of Gene Silencing
VIGS operates through the plant's post-transcriptional gene silencing (PTGS) pathway, an evolutionarily conserved RNA degradation mechanism. The process begins when a recombinant viral vector introduces target gene sequences into the host plant. During viral replication, double-stranded RNA (dsRNA) molecules form, which the plant recognizes as foreign. These dsRNA intermediates are cleaved by Dicer-like (DCL) enzymes into small interfering RNAs (siRNAs) 21-24 nucleotides in length. These siRNAs are then incorporated into an RNA-induced silencing complex (RISC), where they serve as guides to direct the sequence-specific degradation of complementary endogenous mRNA transcripts [79] [3]. The entire process represents a sophisticated cellular defense mechanism that has been co-opted as a powerful functional genomics tool.
Table 1: Key Components of the VIGS Molecular Machinery
| Component | Function | Role in VIGS |
|---|---|---|
| Viral Vector | Delivers target gene fragment into plant cells | Serves as vehicle for inserting host gene sequences |
| Dicer-like (DCL) Enzymes | Processes dsRNA into small RNAs | Cleaves viral dsRNA into siRNAs |
| Small Interfering RNAs (siRNAs) | Guide sequence-specific silencing | Direct RISC to complementary mRNA targets |
| RNA-Induced Silencing Complex (RISC) | Executes mRNA cleavage | Degrades target mRNAs through argonaute proteins |
Visualization of VIGS Workflow and Molecular Mechanism
The following diagram illustrates the integrated experimental workflow and molecular mechanism of VIGS:
Figure 1: Integrated VIGS experimental workflow and molecular mechanism. The dashed lines connect corresponding stages between the practical workflow and cellular processes.
Direct Comparison: VIGS Versus Stable Transformation Approaches
Key Parameter Comparisons Across Plant Species
The practical advantages of VIGS become evident when directly comparing its performance metrics with stable transformation techniques across multiple parameters and plant systems.
Table 2: VIGS vs. Stable Transformation: Comparative Performance Across Plant Species
| Parameter | VIGS | Stable Transformation | Experimental Evidence |
|---|---|---|---|
| Time Requirement | 2-4 weeks for phenotypic analysis [50] [78] | 3-9 months for transgenic line generation | Tea plants: VIGS phenotypes in 12-25 days [78] |
| Silencing Efficiency | 65-95% in soybean [77]; 83% in Styrax japonicus [18] | Highly variable; dependent on transformation efficiency | Soybean TRV system: 65-95% efficiency range [77] |
| Technical Requirements | Agrobacterium-mediated infiltration; no tissue culture needed [4] [78] | Specialized tissue culture, selection, and regeneration systems | Luffa: No transformation system; VIGS established [4] |
| Throughput Capacity | High-throughput; 38 genes simultaneously screened in watermelon [50] | Low to medium throughput; limited by transformation efficiency | Watermelon: 38 male sterility genes screened [50] |
| Applicability to Recalcitrant Species | Successful in tea plants [78], luffa [4], Camellia drupifera [80] | Limited to transformable genotypes | Tea plants: Transformation difficult; VIGS established [78] |
Complementary Strengths and Limitations
While VIGS offers remarkable speed advantages, a balanced perspective acknowledges its limitations compared to stable transformation. VIGS induces transient silencing rather than permanent genetic modification, which means the silencing effect diminishes over time and is not heritable. In contrast, stable transformation creates permanent, heritable changes suitable for long-term studies and breeding programs. Additionally, VIGS can produce mild viral symptoms in some host plants, potentially complicating phenotypic interpretation, though TRV vectors are noted for causing minimal symptoms [77]. The technology also faces challenges in targeting meristematic tissues due to viral exclusion from meristems, potentially limiting its application for studying certain developmental processes [81].
VIGS Implementation: Experimental Designs and Protocols
Case Studies in Crop Plants
Soybean Functional Genomics Using TRV-Based VIGS
Researchers established a tobacco rattle virus (TRV)-based VIGS system in soybean to address limitations of previous bean pod mottle virus (BPMV) systems that often required particle bombardment. The optimized protocol used Agrobacterium tumefaciens GV3101 carrying pTRV1 and pTRV2 vectors delivered through cotyledon node infection [77]. This method achieved high infection efficiency (exceeding 80%, up to 95% for 'Tianlong 1') by bisecting sterilized, pre-swollen soybean seeds and immersing the fresh explants in Agrobacterium suspension for 20-30 minutes. The system successfully silenced key genes including phytoene desaturase (GmPDS)—resulting in photobleaching—as well as the rust resistance gene GmRpp6907 and defense-related GmRPT4, confirming the platform's robustness for studying disease resistance pathways [77].
Watermelon Male Sterility Gene Screening
In watermelon, where stable transformation is extremely challenging, researchers developed a cucumber fruit mottle mosaic virus (CFMMV)-based VIGS vector (pCF93) for high-throughput functional screening. This system screened 38 candidate genes related to male sterility simultaneously, identifying 8 genes that produced male-sterile flowers with abnormal stamens and no pollen when silenced [50]. The vector was introduced into the small watermelon cultivar 'DAH' through Agrobacterium infiltration, enabling large-scale screening in a limited cultivation area. This approach demonstrated how VIGS can rapidly triage candidate genes before committing to more resource-intensive stable transformation approaches for the most promising targets.
Successful implementation of VIGS technology requires specific biological materials and optimization of key parameters. The following table summarizes critical components for establishing VIGS in plant systems.
Table 3: Essential Research Reagents for VIGS Implementation
| Reagent/Resource | Function/Purpose | Examples & Optimization Notes |
|---|---|---|
| Viral Vectors | Deliver target gene fragments; trigger silencing | TRV (broad host range) [77], CFMMV (cucurbits) [50], CGMMV (luffa) [4] |
| Agrobacterium Strains | Mediate vector delivery into plant cells | GV3101 most common; optical density (OD600 = 0.5-1.0) critical [18] [4] |
| Infiltration Buffers | Enhance Agrobacterium infection efficiency | Typically contain MgCl₂, MES, acetosyringone (150-200 μM) [18] [80] |
| Marker Genes | Visual validation of silencing efficiency | PDS (photobleaching) [77] [4], POR1 (chlorophyll deficiency) [78] |
| Target Gene Fragments | Sequence-specific silencing | 200-300 bp fragments with specific design considerations [50] [80] |
Integration Strategy: Positioning VIGS in the Functional Genomics Pipeline
The most effective application of VIGS positions it as a screening and prioritization tool within a broader functional genomics workflow rather than as a replacement for stable transformation. An integrated approach might begin with genome-wide or transcriptome-based identification of candidate genes, followed by VIGS-based rapid screening to identify those yielding phenotypes of interest. The most promising candidates would then advance to stable transformation for comprehensive validation and breeding applications. This tiered strategy maximizes efficiency by focusing extensive stable transformation efforts only on genes with the highest potential value.
This integrated approach is particularly valuable for studying complex traits controlled by multiple genes. For example, in the watermelon male sterility study [50], VIGS enabled researchers to rapidly narrow 38 candidates to 8 high-priority targets—a 79% reduction in candidates requiring further investigation. Similarly, in soybean [77], VIGS provided rapid functional validation of disease resistance genes before committing to lengthy breeding programs. The technology also shows promise for inducing heritable epigenetic modifications through RNA-directed DNA methylation, potentially expanding its applications beyond transient silencing [79].
Future Perspectives and Concluding Remarks
VIGS technology continues to evolve with emerging enhancements including virus-induced genome editing (VIGE) and virus-induced overexpression (VOX) that further expand its utility [4]. The development of new viral vectors with improved host ranges and silencing efficiency addresses current limitations. When integrated with multi-omics technologies, VIGS creates a powerful platform for accelerating gene function discovery in plants.
In conclusion, VIGS serves as an indispensable rapid screening tool that complements rather than replaces stable transformation approaches. Its unique advantages in speed, cost-effectiveness, and applicability to recalcitrant species make it particularly valuable for preliminary gene function assessment and candidate gene prioritization. By implementing VIGS as an initial screening step, researchers can optimize resource allocation, focus stable transformation efforts on the most promising candidates, and dramatically accelerate the pace of functional genomics research in both model and crop plants.
In the field of plant functional genomics, establishing a direct link between a gene and its resulting phenotype is a fundamental objective. Two powerful technologies, Virus-Induced Gene Silencing (VIGS) and CRISPR/Cas9 genome editing, have emerged as cornerstone methods for this purpose. While each approach functions through distinct molecular mechanisms—triggering transient transcript knockdown versus creating permanent DNA mutations—they offer complementary strengths that can be strategically integrated within a single research workflow. This guide provides a comparative analysis of VIGS and CRISPR/Cas9, presenting experimental data and protocols that enable researchers to leverage both methodologies for robust, high-throughput validation of plant gene function.
VIGS is a transient assay that harnesses the plant's RNA interference (RNAi) machinery. Researchers use recombinant viral vectors to deliver host-derived gene fragments, which trigger sequence-specific degradation of complementary endogenous mRNA, leading to a temporary knockdown phenotype [3]. CRISPR/Cas9 editing creates permanent, heritable changes to the plant's DNA sequence. The Cas9 nuclease, guided by a custom-designed RNA molecule, induces double-strand breaks at specific genomic loci. These breaks are repaired by the plant's cellular repair machinery, often resulting in insertion or deletion mutations (indels) that disrupt the function of the target gene [82] [83].
Table 1: Fundamental Characteristics of VIGS and CRISPR/Cas9
| Feature | Virus-Induced Gene Silencing (VIGS) | CRISPR/Cas9 Editing |
|---|---|---|
| Molecular Mechanism | Post-transcriptional gene silencing (PTGS) via siRNA-mediated mRNA degradation [3] | Nuclease-induced DNA double-strand break followed by imperfect repair (NHEJ) [82] [83] |
| Nature of Modification | Transient knockdown of transcript levels | Permanent, heritable gene knockout or mutation |
| Temporal Resolution | Relatively fast (days to weeks) | Slower, requires stable line generation (weeks to months) [21] |
| Typical Workflow | Agroinfiltration or direct inoculation of viral vector | Stable transformation via Agrobacterium or biolistics; can be transient [21] [84] |
| Primary Application | High-throughput functional screening, preliminary gene validation [3] | Definitive gene function confirmation, trait improvement, generating stable lines [84] [83] |
Quantitative Performance Comparison
The practical performance of VIGS and CRISPR/Cas9 varies significantly across critical metrics such as efficiency, throughput, and mutational load. The following table synthesizes experimental data from recent studies to facilitate direct comparison.
Table 2: Experimental Performance Metrics for VIGS and CRISPR/Cas9
| Performance Metric | VIGS | CRISPR/Cas9 |
|---|---|---|
| Typical Efficiency | High (e.g., 100% yellowing phenotype in N. benthamiana with vCHLI) [85] | Variable, species-dependent (e.g., 18% editing in Fraxinus mandshurica growing points) [84] |
| Throughput Potential | Very High (amenable to rapid, large-scale screening) [85] [3] | Moderate (requires regeneration, screening of multiple lines) [84] |
| Mutational Load | Transient, no genomic change | Stable, defined indels or edits at target locus |
| Phenotype Penetrance | Can be variable, dose-dependent [3] | High in homozygous, stable lines |
| Multiplexing Capacity | Limited by viral vector capacity | High (multiple gRNAs can target several genes simultaneously) [86] [83] |
| Optimal Use Case | Rapid phenotype assessment, functional redundancy analysis (e.g., in polyploids) [85] | Detailed functional studies, trait introgression, studying lethal genes in heterozygotes |
A key advancement in VIGS is the development of virus-delivered short RNA inserts (vsRNAi). This approach uses inserts as short as 20-32 nucleotides, nearly 10-fold smaller than those used in conventional VIGS, which simplifies vector engineering and improves specificity by minimizing off-target effects [85]. Studies in Nicotiana benthamiana demonstrated that vsRNAi as short as 24 nt could effectively produce phenotypic alterations, such as leaf yellowing from silencing the CHLI gene, with a significant reduction in chlorophyll levels [85].
Integrated Experimental Workflows
The sequential application of VIGS for initial screening followed by CRISPR/Cas9 for definitive validation constitutes a powerful strategy for confirming plant gene function. This integrated approach maximizes throughput while ensuring rigorous, conclusive results.
Workflow Description and Case Study
The integrated workflow begins with the identification of candidate genes, often from omics studies. VIGS is then deployed for high-throughput, transient knockdown of these candidates, allowing for rapid phenotypic assessment. Promising hits from this initial screen are advanced to CRISPR/Cas9 for the generation of stable, heritable mutations, which provide definitive confirmation of the gene's function [3] [83].
Case Study: Functional Analysis in Polyploid Species Polyploid plants like Nicotiana benthamiana possess complex genomes with functionally redundant homeologous gene pairs, which can obscure phenotypes in single-gene knockout studies. In one study, researchers first used a TRV-based VIGS vector (pLX-TRV2) carrying a 32-nt vsRNAi insert targeting a conserved region of the CHLI gene. This resulted in a systemic leaf yellowing phenotype and a significant reduction in chlorophyll levels within 10 days, effectively silencing both homeologs simultaneously [85]. This rapid VIGS-based confirmation of gene function could be followed by CRISPR/Cas9 to create stable mutant lines, using a multi-gene CRISPR mutagenesis approach to tackle the inherent functional redundancy [85].
Essential Research Reagent Solutions
The successful implementation of these technologies relies on a toolkit of specialized biological reagents and vectors. The following table catalogues key solutions for setting up VIGS and CRISPR/Cas9 experiments.
Table 3: Key Research Reagent Solutions for VIGS and CRISPR/Cas9
| Reagent / Solution | Function / Description | Example Use Case |
|---|---|---|
| JoinTRV System (pLX-TRV1/pLX-TRV2) | A bipartite VIGS vector system based on Tobacco Rattle Virus (TRV) [85] [3] | Systemic silencing in solanaceous plants (e.g., N. benthamiana, tomato) [85] |
| vsRNAi Oligonucleotides | Short, custom-synthesized DNA pairs (20-32 nt) for targeting specific genes [85] | High-specificity silencing with minimal off-target effects; cloned into TRV2 [85] |
| Agrobacterium Strains (e.g., GV3101) | Delivery vehicle for viral vectors or CRISPR constructs via agroinfiltration [21] | Transient transformation of plant leaves for VIGS or editing [21] |
| CRISPR/Cas9 Binary Vectors (e.g., pYLCRISPR/Cas9P35S-N) | Plant transformation vectors harboring Cas9 and sgRNA expression cassettes [84] | Stable plant transformation for generating gene knockouts [84] |
| Viral Suppressors of RNAi (VSRs) | Proteins like P19 or HC-Pro that enhance VIGS by inhibiting host silencing [22] [3] | Co-expression to boost VIGS efficiency and stability in certain hosts [3] |
Detailed Experimental Protocols
Protocol: VIGS using the TRV System and vsRNAi Inserts
This protocol is adapted from studies demonstrating effective silencing in N. benthamiana, tomato, and scarlet eggplant [85].
- vsRNAi Insert Design and Cloning: Design DNA oligonucleotide pairs spanning the 20-32 nt target sequence conserved in the gene of interest. Insert these into the multiple cloning site of the pLX-TRV2 vector using a one-step digestion-ligation reaction [85].
- Agrobacterium Preparation: Transform the recombinant TRV2 vector and the helper TRV1 vector into Agrobacterium tumefaciens (e.g., strain GV3101). Grow individual colonies in liquid culture with appropriate antibiotics until the OD₆₀₀ reaches approximately 0.5. Centrifuge the cultures and resuspend the pellets in an infiltration buffer (10 mM MgCl₂, 10 mM MES, pH 5.6, and 150 µM acetosyringone) [85] [21].
- Plant Infiltration: Mix the TRV1 and TRV2 agrobacterial suspensions in a 1:1 ratio. Using a needleless syringe, pressure-infiltrate the mixture into the abaxial side of fully expanded leaves of young plants (e.g., 3-5 week-old N. benthamiana).
- Phenotype Monitoring: Maintain plants under standard growth conditions. Silencing phenotypes, such as leaf yellowing for the CHLI gene, typically become apparent in systemic (uninoculated) leaves within 10-21 days post-inoculation [85].
- Validation: Confirm silencing efficiency via RT-qPCR to measure transcript abundance of the target gene and, if possible, quantify relevant physiological changes (e.g., chlorophyll fluorometry) [85].
Protocol: CRISPR/Cas9 Editing via Growth Point Transformation
This protocol is based on a system established for the challenging woody species Fraxinus mandshurica and can be adapted for other plants with difficult transformation systems [84].
- Vector Construction: Select three specific knockout targets within the exon of your target gene using online design tools. Clone the synthesized oligonucleotides into a BsaI-digested CRISPR/Cas9 binary vector (e.g., pYLCRISPR/Cas9P35S-N) behind a U6 promoter and transform this vector into Agrobacterium strain EHA105 [84].
- Plant Material Preparation: Use sterile plantlets or embryos germinated on solid medium for a short period (e.g., 7 days).
- Agro-infection of Growth Points: Grow the Agrobacterium culture to an OD₆₀₀ of 0.5-0.8. Resuspend the bacterial pellet in a transformation solution (e.g., containing MES, CaCl₂, acetosyringone, sucrose, mannitol, and DTT). Immerse the plant growth points in this suspension for a defined period (e.g., 30 minutes) [84].
- Co-cultivation and Selection: Blot-dry the infected growth points and co-cultivate them on solid medium for several days. Subsequently, transfer them to a selection medium containing antibiotics (e.g., kanamycin) to inhibit Agrobacterium overgrowth and select for transformed plant cells.
- Regeneration and Screening: Induce and cultivate clustered buds on selective media. Extract genomic DNA from the regenerated shoots and use PCR-based methods to detect mutations at the target locus. The edited, chimeric plants can then be used to induce and screen for homozygous mutants [84].
VIGS and CRISPR/Cas9 are not mutually exclusive technologies but rather complementary pillars in the plant functional genomics toolkit. VIGS offers an unparalleled platform for speed and throughput, ideal for initial screening and testing hypotheses in a transient context. CRISPR/Cas9 provides the definitive, stable genetic evidence required for conclusive validation and trait development. By strategically integrating these approaches—using VIGS as a rapid filter to prioritize candidates for subsequent CRISPR/Cas9 editing—researchers can construct an efficient and robust pipeline for discovering and validating gene functions, ultimately accelerating crop improvement programs.
In plant functional genomics, unraveling gene function is pivotal for understanding the genetic control of agronomically valuable traits. Virus-Induced Gene Silencing (VIGS) has emerged as a powerful reverse genetics tool that enables transient, sequence-specific downregulation of target genes by harnessing the plant's innate RNA interference machinery [79] [3]. Unlike stable transformation techniques, VIGS offers rapid (1-2 weeks from infection to silencing) characterization of gene function without the need for developing stable transformants, making it particularly valuable for high-throughput functional screening in species recalcitrant to transformation [87] [3]. However, the transient and potentially incomplete nature of VIGS-mediated knockdown necessitates rigorous validation through multi-omics approaches, especially integration with transcriptomic data, to confirm target gene suppression and identify downstream consequences [79] [88].
Multi-omics integration represents a paradigm shift in biological research, enabling researchers to move beyond single-layer analyses to obtain a holistic view of molecular processes [89]. By combining data from genomics, transcriptomics, epigenomics, and other molecular levels, scientists can assess the flow of information from genotype to phenotype and decipher complex regulatory networks [89] [88]. In the context of VIGS studies, multi-omics correlation not only validates silencing efficiency but also reveals compensatory mechanisms, off-target effects, and the broader transcriptional rewiring that occurs in response to targeted gene suppression [79] [88]. This integrated approach is transforming how researchers validate gene function in plant systems, particularly for traits with complex genetic architecture such as flowering time, stress responses, and unique metabolic pathways [88] [3].
Multi-Omics Integration Methodologies in Functional Genomics
Technical Approaches for Data Integration
The integration of silencing data with transcriptomics employs diverse computational strategies that can be broadly categorized into simultaneous, sequential, and network-based approaches. Simultaneous integration methods analyze multiple omics datasets in parallel, preserving the inter-relationships between different molecular layers. These include multiple kernel learning, matrix factorization, and multivariate statistical methods that simultaneously decompose multi-omics data into latent components [89]. For instance, in the study of Arabidopsis traits, researchers have successfully employed ridge regression Best Linear Unbiased Prediction (rrBLUP) and Random Forest algorithms to integrate genomic, transcriptomic, and methylomic data for improved trait prediction [88].
Sequential integration methods analyze omics data in a stepwise manner, where the output from one analysis informs the next. A prominent example is SIMO (Spatial Integration of Multi-Omics), a computational tool designed specifically for spatial integration of single-cell multi-omics data [90]. SIMO employs a sequential mapping process that begins with integrating spatial transcriptomics with single-cell RNA-seq data, followed by mapping of non-transcriptomic single-cell data such as chromatin accessibility and DNA methylation through probabilistic alignment. This approach uses the k-nearest neighbor (k-NN) algorithm to construct spatial graphs and modality maps, calculating mapping relationships through fused Gromov-Wasserstein optimal transport [90].
Network-based integration methods construct biological networks that connect different omics layers through functional relationships. These approaches often use gene co-expression networks derived from transcriptomics data as a scaffold for integrating other molecular data types [91]. For example, a novel automated function prediction method for Arabidopsis thaliana leverages complementary information from multiple expression datasets by analyzing study-specific gene co-expression networks, outperforming state-of-the-art expression-based approaches [91]. This method successfully assigned validated biological process annotations to 42.6% of previously unknown genes in Arabidopsis, demonstrating the power of network-based multi-omics integration for gene function discovery [91].
Workflow for Integrating VIGS Data with Transcriptomics
The following diagram illustrates the comprehensive workflow for correlating VIGS silencing data with transcriptomic profiles, from experimental design through biological insight:
This integrated workflow ensures that VIGS-mediated silencing phenotypes can be rigorously correlated with transcriptomic changes, providing a comprehensive understanding of gene function within broader biological networks.
Comparative Analysis of Gene Silencing and Editing Technologies
RNAi/VIGS vs. CRISPR: Mechanisms and Applications
Understanding the relative strengths and limitations of different gene perturbation technologies is essential for appropriate experimental design in functional genomics. The following table compares the key characteristics of RNAi/VIGS and CRISPR-based approaches:
Table 1: Comparison of Gene Silencing and Editing Technologies
| Feature | RNAi/VIGS | CRISPR Knockout | CRISPR Interference (CRISPRi) |
|---|---|---|---|
| Mechanism of Action | Post-transcriptional mRNA degradation or translational inhibition [38] [92] | DNA double-strand breaks leading to frameshift mutations [38] [92] | Catalytically dead Cas9 blocks transcription [92] [93] |
| Level of Intervention | mRNA level (cytoplasmic) [38] [92] | DNA level (nuclear) [38] [93] | DNA level (transcriptional regulation) [92] |
| Persistence of Effect | Transient (knockdown) [38] [93] | Permanent (knockout) [38] [93] | Reversible (knockdown) [92] |
| Efficiency | Variable; incomplete knockdown common [93] | High; complete knockout possible [93] | Variable; depends on target site [92] |
| Off-Target Effects | High; sequence-dependent and independent effects [38] [92] | Moderate; improved with high-fidelity Cas variants [38] [93] | Lower; no DNA cleavage [92] |
| Applications in Functional Genomics | Rapid screening, essential gene study, dose-response studies [87] [93] | Complete loss-of-function, stable line generation [38] [92] | Temporal control, essential gene study [92] |
| Therapeutic Relevance | Better representation of pharmacological inhibition [93] | Limited for transient modulation | Potential for controlled regulation [92] |
Multi-Omics Performance Metrics Across Technologies
The integration of multi-omics data has revealed distinct performance characteristics across different gene perturbation technologies. Recent studies in Arabidopsis thaliana provide quantitative insights into how different omics data types contribute to trait prediction and functional annotation:
Table 2: Multi-Omics Performance in Functional Genomics Studies
| Omics Data Type | Prediction Accuracy (PCC) | Key Strengths | Limitations |
|---|---|---|---|
| Genomic (G) | 0.15-0.65 (flowering time) [88] | Direct representation of genetic variation; stable measurement [88] | Limited functional interpretation; distal to phenotype [88] |
| Transcriptomic (T) | 0.20-0.63 (flowering time) [88] | Functional readout of cellular state; direct pathway information [88] | Context-dependent; technical variability [89] [88] |
| Methylomic (M) | 0.18-0.60 (flowering time) [88] | Epigenetic regulation capture; environmental response imprint [79] [88] | Confounding with genetic variation; complex data structure [88] |
| Integrated Multi-Omics | 0.25-0.72 (flowering time) [88] | Superior predictive power; comprehensive biological view [89] [88] | Computational complexity; data integration challenges [89] [88] |
These quantitative comparisons demonstrate that while individual omics data types provide valuable insights, integrated multi-omics approaches consistently outperform single-omics analyses in predicting complex traits and annotating gene function [88]. The performance gains are particularly evident for complex traits such as flowering time, where integrated models achieved Pearson Correlation Coefficients (PCC) of up to 0.72 compared to maximum PCCs of 0.65, 0.63, and 0.60 for genomic, transcriptomic, and methylomic models alone, respectively [88].
Experimental Protocols and Validation Frameworks
VIGS-Transcriptomics Integration Protocol
A robust experimental protocol for correlating VIGS silencing data with transcriptomic profiles involves standardized steps from vector construction to multi-omics integration:
VIGS Vector Construction and Plant Inoculation:
- Select appropriate viral vector (TRV-based systems most common for Solanaceae) and design insert sequences targeting genes of interest (300-500 bp with 40-80% GC content) [3].
- Clone target fragments into TRV2 vector while maintaining TRV1 for viral replication and movement proteins [87] [3].
- Inoculate plants via agroinfiltration (OD600 = 0.3-1.0) at 4-6 leaf stage, ensuring proper controls including empty vector and non-silenced plants [3].
- Maintain plants under optimized conditions (temperature: 19-22°C; photoperiod: 10-12 hours; humidity: 60-80%) to maximize silencing efficiency [3].
Efficiency Validation and RNA Sequencing:
- Confirm silencing efficiency 1-3 weeks post-inoculation using qRT-PCR (target: 70-95% reduction in mRNA) and document phenotypic consequences [3].
- Extract high-quality RNA (RIN > 8.0) from silenced and control tissues, with biological replicates (n ≥ 4) [88].
- Prepare stranded mRNA sequencing libraries and sequence on appropriate platform (Illumina recommended: 30-50 million reads/sample, 150 bp paired-end) [88].
Transcriptomic Data Analysis and Integration:
- Process raw reads: quality control (FastQC), adapter trimming, and alignment to reference genome (STAR/Hisat2) [88] [91].
- Perform differential expression analysis (DESeq2/edgeR) with false discovery rate correction (FDR < 0.05) [88].
- Conduct co-expression network analysis (WGCNA) and pathway enrichment (GO, KEGG) to identify affected biological processes [91].
- Integrate with additional omics data (methylation, proteomics) using tools such as SIMO for spatial context or network-based integration for functional annotation [90] [91].
Research Reagent Solutions Toolkit
Successful integration of VIGS with multi-omics approaches requires specific research reagents and computational tools:
Table 3: Essential Research Reagents and Tools for VIGS-Transcriptomics Integration
| Reagent/Tool | Function | Application Notes |
|---|---|---|
| TRV-Based VIGS Vectors | Delivery of target gene sequences for silencing [87] [3] | Bipartite system (TRV1/TRV2); broad host range; efficient in meristems [3] |
| Agrobacterium tumefaciens | Bacterial delivery system for viral vectors [3] | Strain GV3101 most common; optical density critical for efficiency [3] |
| RNA Extraction Kits | High-quality RNA isolation for transcriptomics | Must yield RIN > 8.0; compatible with challenging plant tissues |
| RNA-seq Library Prep Kits | cDNA library construction for sequencing | Stranded mRNA protocols recommended; include rRNA depletion |
| SIMO | Spatial integration of multi-omics single-cell data [90] | Enables mapping of scRNA-seq, scATAC-seq to spatial contexts [90] |
| Co-expression Network Tools | Construction of gene regulatory networks [91] | Study-specific networks improve functional prediction [91] |
| Multi-omics Databases | Reference data for validation and comparison | TCGA, ICGC, CPTAC for cancer; CCLE for cell lines [89] |
Case Studies in Plant Functional Genomics
Arabidopsis Flowering Time Regulation
A comprehensive study integrating multi-omics data for complex trait prediction in Arabidopsis thaliana demonstrated the power of combining genomic, transcriptomic, and methylomic data [88]. Researchers built machine learning models using rrBLUP and Random Forest algorithms to predict six complex traits, including flowering time, from different omics data types individually and in combination [88]. The study revealed several key insights:
- Models based on different omics types (G, T, M) showed comparable prediction accuracy for flowering time, but identified distinct sets of important genes, suggesting complementary biological information [88].
- The correlation between feature importance scores from models built using different omics data was weak (Spearman's ρ: -0.07–0.11), with little overlap of important genes between models [88].
- Integrated models leveraging all three omics data types simultaneously achieved the highest prediction accuracy (PCC up to 0.72 for flowering time) and revealed known and novel gene interactions, extending knowledge about existing regulatory networks [88].
- Nine additional genes identified as important for flowering time from the integrated models were experimentally validated as genuine flowering time regulators, demonstrating the predictive power of multi-omics integration [88].
Network-Based Functional Annotation of Unknown Genes
A novel automated function prediction method leveraging multi-omics networks successfully assigned biological process annotations to previously uncharacterized Arabidopsis genes [91]. This approach:
- Outperformed state-of-the-art expression-based methods by analyzing study-specific gene co-expression networks derived from multiple expression datasets [91].
- Assigned at least one validated annotation to 5,054 (42.6%) previously unknown genes, and at least one novel validated function to 3,408 (53.0%) genes with only computational annotations [91].
- Provided insights into diverse biological processes, including flower and root development, defense responses to pathogens, and phytohormone signaling [91].
- Demonstrated how context-specific networks, modeling seed development and response to water deprivation, could elucidate how previously uncharacterized genes function within biological networks [91].
The integration of VIGS silencing data with transcriptomics and other omics layers represents a powerful paradigm for validating gene function in plant systems. Multi-omics correlation not only confirms target gene knockdown but also reveals the broader transcriptional consequences of gene silencing, including compensatory mechanisms, off-target effects, and pathway-level rewiring [79] [88]. The complementary nature of different omics data types—with transcriptomic, genomic, and methylomic models identifying distinct gene sets despite similar prediction accuracy—highlights the importance of integrated approaches for comprehensive biological understanding [88].
As multi-omics technologies continue to advance, emerging computational tools like SIMO for spatial integration [90] and network-based functional annotation methods [91] are poised to further enhance our ability to extract biological insights from complex datasets. For researchers validating plant gene function through VIGS and knockout studies, the strategic integration of multi-omics approaches provides a robust framework for connecting gene silencing phenotypes to molecular mechanisms, accelerating the discovery of genes controlling agronomically valuable traits and advancing crop improvement efforts [88] [3] [91].
Virus-induced genome editing (VIGE) represents a transformative approach in plant functional genomics, combining the high efficiency of viral vector delivery with the precision of CRISPR/Cas-based editing systems. This technology enables researchers to overcome traditional limitations in genetic transformation, particularly for recalcitrant crop species, while producing transgene-free edited plants in a single generation. As global deregulation of transgene-free genome-edited plants expands, with over 20 countries having established streamlined approval processes, VIGE platforms offer an unprecedented opportunity for accelerated crop improvement. This review provides a comprehensive comparison of current VIGE systems, detailing their molecular mechanisms, performance metrics across plant species, and integration with high-throughput validation platforms. We present experimental protocols and quantitative data to guide researchers in selecting appropriate viral vectors for specific applications, with particular emphasis on overcoming abiotic and biotic stress tolerance in crop plants.
Virus-induced genome editing has emerged as a powerful alternative to stable genetic transformation, addressing critical bottlenecks in plant functional genomics and crop improvement programs. VIGE utilizes engineered viral vectors to deliver CRISPR/Cas components directly to plant cells, enabling targeted mutagenesis without the need for tissue culture or the integration of foreign DNA into the plant genome [22] [94]. This technology builds upon the well-established framework of virus-induced gene silencing (VIGS) but expands its capabilities to include precise genome modifications through the CRISPR/Cas system.
The agricultural imperative driving VIGE development is substantial – plant diseases alone cause estimated yield losses of 20-40% in major global crops such as rice, maize, wheat, potato, and soybean, while abiotic stresses like drought and salinity further reduce potential yields by 20-50% [94]. Traditional breeding methods struggle to address these challenges with sufficient speed, particularly for complex traits controlled by multiple genes. While CRISPR/Cas technology itself has revolutionized plant genome engineering, its delivery through Agrobacterium-mediated transformation or biolistics remains inefficient for many crop species and desirable cultivars [94] [50].
VIGE addresses these limitations by leveraging the natural ability of viruses to systemically infect plants and replicate within cells, effectively amplifying the delivery of editing components throughout plant tissues [22]. The transient nature of viral infection means that editing can occur without permanent integration of viral sequences, potentially yielding transgene-free edited plants in a single generation – a significant regulatory advantage as countries worldwide move to deregulate such plants [22]. This positions VIGE as an essential tool for high-throughput functional genomics and rapid crop improvement, particularly for species resistant to conventional transformation methods.
Molecular Mechanisms and Vector Systems in VIGE
Fundamental Principles of VIGE
The molecular foundation of VIGE combines the efficient delivery capabilities of plant viruses with the precision targeting of CRISPR/Cas systems. When a modified viral vector carrying CRISPR components infects a plant cell, it hijacks the host's replication machinery to amplify these components, simultaneously spreading systemically throughout the plant. The Cas nuclease (most commonly Cas9) creates double-strand breaks at target genomic loci guided by virus-derived gRNAs, triggering the plant's endogenous DNA repair mechanisms that introduce targeted mutations through error-prone non-homologous end joining (NHEJ) or, less frequently, precise edits via homology-directed repair (HDR) when donor templates are present [94].
Unlike stable transformation, where CRISPR components integrate into the plant genome, VIGE maintains these components episomally within viral particles. This transient presence significantly reduces the likelihood of persistent transgenes, addressing regulatory concerns associated with genetically modified organisms [22]. The system leverages the virus's natural life cycle while subverting it for genome engineering purposes, creating a powerful synergy between virology and genome editing technologies.
Comparative Analysis of Viral Vector Systems
VIGE implementation varies significantly depending on the viral vector system employed, with each offering distinct advantages and limitations based on genome type, cargo capacity, and host range. The table below provides a systematic comparison of major viral vectors used in VIGE applications:
Table 1: Comparison of Viral Vector Systems for VIGE Applications
| Virus Type | Representative Vectors | Cargo Capacity | Editing Efficiency | Host Range | Key Limitations |
|---|---|---|---|---|---|
| RNA Viruses | Tobacco Rattle Virus (TRV), Barley Stripe Mosaic Virus (BSMV) | Low to Moderate (~1.5 kb) | Variable (5-30%) | Broad (TRV) to Monocot-specific (BSMV) | Limited cargo capacity, plant immune responses |
| DNA Viruses | Geminiviruses (Bean Yellow Dwarf Virus) | Moderate (~2.5 kb) | Moderate to High (up to 40% in some systems) | Primarily dicots | Small genome size, host specificity |
| Rhabdoviruses | Sonchus Yellow Net Rhabdovirus (SYNV) | High (up to 5 kb) | High (demonstrated in N. benthamiana) | Narrow | Complex vector engineering, limited host range |
| Tobamoviruses | Cucumber Fruit Mottle Mosaic Virus (CFMMV), Cucumber Green Mottle Mosaic Virus (CGMMV) | Low to Moderate | Established for VIGS, VIGE emerging | Cucurbit-specific | Meristem exclusion, tissue specificity |
The selection of an appropriate viral vector represents a critical decision point in VIGE experimental design, requiring careful consideration of the target plant species, tissue specificity requirements, and the size of CRISPR components needed. RNA viruses like TRV have been widely adopted due to their broad host range and established VIGS protocols [4] [81], while DNA viruses such as geminiviruses offer the advantage of replication in the nucleus where CRISPR/Cas editing occurs [22]. Larger viruses like SYNV can accommodate substantial genetic cargo, including multiple gRNAs or even base editing systems, but present greater challenges in vector engineering and host range limitations [94].
Visualizing the VIGE Workflow and Molecular Mechanism
The following diagram illustrates the fundamental molecular process of virus-induced genome editing in plant cells:
Figure 1: Molecular Mechanism of VIGE in Plant Cells
Beyond the core mechanism, the practical implementation of VIGE requires a standardized experimental workflow that spans from vector design to validation of edited plants:
Figure 2: Standard VIGE Experimental Workflow
High-Throughput Validation Platforms for VIGE
Integration of VIGE with Functional Genomics Platforms
The true potential of VIGE emerges when integrated with high-throughput validation platforms that enable rapid screening and characterization of edited lines. Next-generation sequencing technologies provide the foundation for these pipelines, with whole exome sequencing (WES) platforms offering cost-effective validation of editing outcomes across multiple samples simultaneously [28]. Recent comparative studies of commercial exome capture platforms – including TargetCap (BOKE), xGen (IDT), EXome (Nanodigmbio), and Twist – have demonstrated their utility in variant detection following genome editing applications [28].
Quantitative PCR platforms represent another essential component of the VIGE validation pipeline, particularly for assessing the efficiency of viral delivery and editing component expression. Comparative analyses of high-throughput qPCR systems, including the OpenArray (Life Technologies) and Dynamic Array (Fluidigm) platforms, have revealed critical performance differences relevant to VIGE validation [95]. The standard 96-well platform (ViiA7) demonstrated the lowest coefficient of variation (median CV of 0.6%) and highest fidelity (99.23% of replicates differing by <1 CT value), while ultra-high-throughput systems showed increased variability, particularly for low-abundance transcripts [95]. These technical considerations directly impact the reliability of editing efficiency assessments in VIGE experiments.
Performance Metrics Across Validation Platforms
The selection of appropriate validation platforms requires careful consideration of performance metrics specific to VIGE applications. The following table compares key platform characteristics based on empirical data:
Table 2: Performance Comparison of High-Throughput Validation Platforms for VIGE
| Platform | Throughput | Reaction Volume | Sensitivity (Low Copy Detection) | Replicate Variability (CV) | Best Application in VIGE Pipeline |
|---|---|---|---|---|---|
| Standard 96-well (ViiA7) | 96 samples/run | 5 μl | High | 0.6% (median) | Gold standard for validation of key edits |
| TaqMan Low Density Array (TLDA) | 384 samples/run | 1 μl | Moderate | 8.3% (median) | Medium-throughput screening |
| OpenArray | 3072 samples/run | 33 nl | Moderate to Low | 2.1% (median) | High-throughput initial screening |
| Dynamic Array | 9216 samples/run | 15 nl | Lowest | 9.5% (median) | Ultra-high-throughput applications |
Data derived from platform comparison studies indicates that sensitivity for detecting low-copy number targets is inversely related to reaction volume, with nanoliter-scale systems like the Dynamic Array showing reduced performance for rare targets [95]. This has direct implications for VIGE validation, where early detection of editing events may involve low-abundance molecules. Platform selection must therefore balance throughput requirements with analytical sensitivity based on the specific stage of the VIGE pipeline.
Essential Research Reagent Solutions for VIGE
Successful implementation of VIGE technology requires access to specialized reagents and materials optimized for viral vector delivery and genome editing in plants. The following table details key research solutions employed in established VIGE protocols:
Table 3: Essential Research Reagents for VIGE Implementation
| Reagent Category | Specific Examples | Function in VIGE | Implementation Notes |
|---|---|---|---|
| Viral Vectors | TRV, BSMV, CFMMV, CGMMV, SYNV | Delivery of CRISPR components | Selection based on host compatibility and cargo requirements |
| Agrobacterium Strains | GV3101, LBA4404 | Delivery of viral vectors to plants | Standard for agroinfiltration protocols |
| CRISPR Components | Cas9 variants, gRNA scaffolds | Genome editing machinery | Codon-optimization for plant expression |
| Selection Markers | Antibiotic resistance genes | Vector maintenance and selection | Kanamycin, rifampicin common for bacterial components |
| Infiltration Buffers | MgCl₂, MES, acetosyringone | Enhancement of Agrobacterium infection | Critical for efficient vector delivery |
| Silencing Suppressors | HC-Pro, p19 | Inhibition of plant RNAi defense | Enhances viral persistence and editing window |
The selection and optimization of these core reagents directly impact editing efficiency across different plant species. For example, the pCF93 vector derived from cucumber fruit mottle mosaic virus has demonstrated particular effectiveness in cucurbit species, enabling functional analysis of male sterility genes in watermelon through silencing of 8 out of 38 candidate genes [50]. Similarly, tobacco rattle virus (TRV) vectors have shown broad host range compatibility, making them suitable for initial VIGE system development in non-model species [4] [81].
Experimental Protocols for VIGE Implementation
Vector Construction and Plant Inoculation
The implementation of VIGE begins with careful vector design and assembly. For RNA viruses like TRV, the strategy typically involves separating Cas9 and gRNA expression cassettes between different viral components or utilizing subgenomic promoters to coordinate expression [22] [94]. A standardized protocol for VIGE vector construction and plant inoculation includes:
Vector Assembly: Clone target-specific gRNA sequences (typically 150-500 bp) into the viral vector backbone using appropriate restriction sites or recombination-based cloning. For DNA viruses like geminiviruses, the entire Cas9-gRNA expression cassette may be inserted, while for RNA viruses with limited capacity, creative partitioning of components is often necessary [94].
Agrobacterium Preparation: Transform the assembled vector into appropriate Agrobacterium tumefaciens strains (e.g., GV3101). Grow single colonies in selective liquid media (YEP with appropriate antibiotics) overnight at 28°C. Subculture to larger volumes and grow until OD600 reaches 0.6-0.8 [4].
Bacterial Resuspension: Pellet bacterial cultures by centrifugation and resuspend in infiltration buffer (10 mM MgCl₂, 10 mM MES, 200 μM acetosyringone). Adjust the final OD600 to 0.8-1.0 and incubate at room temperature for 2-4 hours to induce virulence gene expression [4].
Plant Inoculation: For seedlings at the 2-4 leaf stage, use a needleless syringe to infiltrate the bacterial suspension through the abaxial side of leaves. Alternatively, mechanical inoculation by rubbing carbonundum-dusted leaves with bacterial suspension can be employed. Maintain inoculated plants under high humidity conditions for 24-48 hours to facilitate infection [4].
Systemic Spread and Editing: Monitor plants for viral symptoms and systemic spread over 1-3 weeks post-inoculation. The editing window typically peaks during active viral replication, with optimal harvest times for validation experiments varying by plant species and viral vector [22].
Validation and Analysis of Editing Events
Robust validation of editing outcomes is essential for interpreting VIGE experiments. A multi-tiered approach to validation provides comprehensive assessment of editing efficiency and specificity:
Molecular Validation of Editing: Extract genomic DNA from systemic leaves showing viral infection symptoms. Amplify target regions by PCR using gene-specific primers and analyze editing efficiency through restriction fragment length polymorphism (RFLP) assays if the edit disrupts a restriction site, or through sequencing-based methods such as T7E1 or GUIDE-seq assays [94].
Assessment of Viral Load and Spread: Quantify viral titer in different plant tissues using RT-qPCR with virus-specific primers. This correlation between viral distribution and editing efficiency informs optimization of harvest timing and tissue selection [95].
Off-Target Analysis: Identify potential off-target sites in the genome using computational prediction tools based on sequence similarity to the intended target. Amplify and sequence these regions to assess editing specificity, particularly when deploying VIGE in translational applications [22].
Phenotypic Validation: For characterized genes, correlate editing outcomes with expected phenotypic changes. Common markers include photobleaching for PDS gene editing or developmental abnormalities for transcription factors involved in growth regulation [4] [96].
Inheritance Analysis: Collect seeds from edited plants and genotype subsequent generations to confirm heritability of edits and assess segregation patterns. This step is crucial for confirming transgene-free editing events [22].
Current Challenges and Future Perspectives
Despite significant advances, VIGE technology faces several persistent challenges that represent active areas of research and development. Key limitations include insufficient viral vector capacity for larger CRISPR systems, plant immune responses that constrain editing windows, meristem exclusion that limits germline editing, and host specificity that restricts vector application across diverse crop species [22]. Current research focuses on addressing these constraints through multiple innovative approaches.
Future directions in VIGE technology development include the creation of viral vectors with expanded cargo capacity through minimalistic design approaches, fusion of mobile RNA elements to enhance meristem penetration, and engineering of tissue-specific promoters to target editing to desired organs [22]. The integration of VIGE with emerging CRISPR technologies such as base editing and prime editing represents another promising frontier, potentially enabling more precise editing outcomes without requiring double-strand breaks. Additionally, the combination of VIGE with high-throughput phenotyping platforms and automated sample processing will further accelerate the functional genomics pipeline.
As regulatory frameworks for genome-edited plants continue to evolve globally, with the United States Department of Agriculture, Argentina, Japan, Australia, Brazil, China, and the UK having established streamlined approval processes for transgene-free edits, VIGE stands positioned to dramatically accelerate crop improvement efforts [22]. The technology's ability to rapidly validate gene function in diverse genetic backgrounds without lengthy backcrossing protocols offers particular promise for addressing pressing agricultural challenges related to climate change, disease resistance, and nutritional enhancement.
The continued refinement of VIGE platforms, coupled with thoughtful integration into comprehensive functional genomics workflows, will undoubtedly transform plant genetic research and precision breeding in the coming years. As vector efficiency improves and host range expands, researchers will increasingly leverage this powerful technology to dissect complex biological pathways and develop improved crop varieties with enhanced productivity and resilience.
Conclusion
VIGS has established itself as an indispensable reverse genetics tool that provides a rapid, cost-effective alternative to traditional stable transformation for gene function validation, particularly in genetically recalcitrant species. When integrated with emerging genome editing technologies like CRISPR/Cas9, VIGS enables powerful combinatorial approaches—from initial high-throughput screening to precise functional characterization. The future of plant functional genomics will be shaped by further optimization of viral vectors, expansion to non-model species, and the development of VIGS-mediated genome editing (VIGE) systems. These advances will accelerate the discovery of agronomically valuable genes and their deployment in precision breeding programs, ultimately contributing to enhanced crop resilience and productivity in the face of global challenges.
References
- 1. Suppress to Survive—Implication of Plant Viruses in PTGS [https://pmc.ncbi.nlm.nih.gov/articles/PMC4432016/]
- 2. Virus-induced gene silencing: A versatile tool for discovery ... [https://www.sciencedirect.com/science/article/abs/pii/S0981942809001934]
- 3. Virus-Induced Gene Silencing (VIGS) in Functional Genomics [https://www.mdpi.com/2311-7524/11/11/1297]
- 4. Establishment of virus-induced gene silencing (VIGS) system ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-023-01064-4]
- 5. Antiviral RNA interference in plants: Increasing complexity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12546453/]
- 6. Suppression of Posttranscriptional Gene Silencing [https://www.sciencedirect.com/science/article/pii/S0092867400816141]
- 7. Efficient Virus-Induced Gene Silencing in Roots Using a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC535832/]
- 8. Establishing a Virus-Induced Gene Silencing System in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10346259/]
- 9. Molecular Bases of Viral RNA Targeting by Viral Small ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1866121/]
- 10. RNA interference and turnover in plants -a complex ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1608888/full]
- 11. Fragile X-related protein and VIG associate with the RNA ... [https://genesdev.cshlp.org/content/16/19/2491.long]
- 12. Establishment of virus-induced gene silencing (VIGS) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10470258/]
- 13. Rethinking de novo genes in plants: mechanisms ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1724832/full]
- 14. Transcriptome-based analysis reveals key molecular ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12305966/]
- 15. Comparative single-cell transcriptomic map reveals ... [https://www.sciencedirect.com/science/article/pii/S2090123225002565]
- 16. Recalcitrance to transformation, a hindrance for genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10551638/]
- 17. Opinion Unlocking in vitro transformation of recalcitrant plants [https://www.sciencedirect.com/science/article/abs/pii/S1360138525001943]
- 18. Styrax japonicus functional genomics: an efficient virus induced gene silencing (VIGS) system - ScienceDirect [https://www.sciencedirect.com/science/article/pii/S2468014123000833]
- 19. A virus-induced gene silencing (VIGS) system for functional genomics in the parasitic plant Striga hermonthica | Plant Methods | Full Text [https://plantmethods.biomedcentral.com/articles/10.1186/1746-4811-10-16]
- 20. Virus-Induced Gene Silencing, a Post Transcriptional Gene Silencing Method - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2699436/]
- 21. Review and Validation of Plant Gene Function Research ... [https://www.mdpi.com/2073-4395/15/3/603]
- 22. Virus-Induced Genome Editing (VIGE): One Step Away ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12111327/]
- 23. In silico prediction of variant effects: promises and limitations ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12304032/]
- 24. Comparative Analysis of Deep Learning Models for ... [https://www.mdpi.com/2073-4425/16/10/1223]
- 25. Simultaneous silencing of two different Arabidopsis genes ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-020-00701-6]
- 26. CRISPR/Cas9-mediated gene knockout screens and target ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5481571/]
- 27. Subtractive genomics and comparative metabolic ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1484423/full]
- 28. Performance comparison of four exome capture platforms on ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-12104-9]
- 29. Establishment of a Virus-Induced Gene-Silencing (VIGS) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9820744/]
- 30. Plant Virus-Based Tools for Studying the Function of Gene ... [https://www.intechopen.com/chapters/1135257]
- 31. A cucumber green mottle mosaic virus vector for virus-induced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6996188/]
- 32. Development of a cucumber green mottle mosaic virus- ... [https://www.sciencedirect.com/science/article/abs/pii/S016609341830332X]
- 33. Establishment of efficient Trichosanthes mottle mosaic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12092892/]
- 34. Genome Characterisation of the CGMMV Virus Population ... [https://www.mdpi.com/1999-4915/15/3/743]
- 35. based virus-induced gene Silencing in Atriplex canescens [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-025-01427-z]
- 36. Engineering PVX vectors harboring heterologous VSRs to ... [https://www.nature.com/articles/s41598-025-24470-1]
- 37. Viral vector-based transient expression systems for plant ... [https://www.frontiersin.org/journals/education/articles/10.3389/feduc.2025.1598673/full]
- 38. RNAi vs. CRISPR: Guide to Selecting the Best Gene ... [https://www.synthego.com/blog/rnai-vs-crispr-guide/]
- 39. Agroinfiltration technique for elucidating the functions of ... [https://www.nature.com/articles/s41598-025-08344-0]
- 40. Establishment of Agrobacterium-Mediated Transient ... [https://www.mdpi.com/2223-7747/14/15/2412]
- 41. An Efficient Agrobacterium-Mediated Transient ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12388925/]
- 42. Vacuum and Co-cultivation Agroinfiltration of (Germinated) ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.00393/full]
- 43. CRISPR/Cas9 based genome editing of Phytoene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11070243/]
- 44. Phytoene Desaturase (PDS) Gene-Derived Markers ... [https://www.mdpi.com/2311-7524/10/3/294]
- 45. Developing a homology-driven gene silencing system in < ... [https://www.maxapress.com/article/id/68c8b5d4fa6c58419c48f3c2]
- 46. Establishment of Transcriptional Gene Silencing Targeting ... [https://pubmed.ncbi.nlm.nih.gov/36437439/]
- 47. Enhancement of Virus-induced Gene Silencing in Tomato ... [https://www.sciencedirect.com/science/article/pii/S1016847823129141]
- 48. Developing and applying a virus-induced gene silencing ... [https://www.sciencedirect.com/science/article/abs/pii/S0378111924009685]
- 49. A Cotyledon-based Virus-Induced Gene Silencing (Cotyledon-VIGS) approach to study specialized metabolism in medicinal plants | Plant Methods | Full Text [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01154-x]
- 50. Virus-induced gene silencing for in planta validation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9706489/]
- 51. Plant non-coding RNAs function in pollen development and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9975556/]
- 52. Current insights and advances into plant male sterility [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1223861/full]
- 53. TaEXPB5 functions as a gene related to pollen ... [https://www.sciencedirect.com/science/article/abs/pii/S0168945222002011]
- 54. Using high-throughput multiple optical phenotyping to ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-021-02377-0]
- 55. Machine Learning for High-Throughput Stress Phenotyping ... [https://www.sciencedirect.com/science/article/pii/S1360138515002630]
- 56. A high-throughput differential chemical genetic screen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12032938/]
- 57. Highly efficient Agrobacterium rhizogenes-mediated ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-023-01078-y]
- 58. Establishment of an Efficient Agrobacterium rhizogenes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12349569/]
- 59. Simplified Protocol to Demonstrate Gene Expression in Nicotiana... [https://bio-protocol.org/en/bpdetail?id=4987&type=0]
- 60. Developing and applying a virus-induced gene silencing ... [https://pubmed.ncbi.nlm.nih.gov/39542283/]
- 61. Enhancing virus-induced gene silencing efficiency in tea ... [https://www.sciencedirect.com/science/article/abs/pii/S0304423824007398]
- 62. An Optimized Protocol to Increase Virus-Induced Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3893464/]
- 63. Establishment of a Virus-Induced Gene-Silencing (VIGS) ... [https://www.mdpi.com/2311-7524/10/4/422]
- 64. Effectiveness of gene silencing induced by viral vectors ... [https://www.sciencedirect.com/science/article/pii/S0042682214001408]
- 65. Grapevine virusA-mediated gene silencing in Nicotiana benthamiana... [https://pubmed.ncbi.nlm.nih.gov/19010356/]
- 66. Genetic architecture of a light-temperature coincidence ... [https://www.nature.com/articles/s41467-025-62194-y]
- 67. Elevated relative humidity significantly decreases ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1678142/full]
- 68. Temperature–Photoperiod Interaction in Rice Phenology for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12238749/]
- 69. Light, Temperature, and Relative Humidity Influence the ... [https://www.hst-j.org/articles/xml/O4qy/]
- 70. A high-throughput virus-induced gene silencing protocol ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3879149/]
- 71. Virus and Viroid-Derived Small RNAs as Modulators ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7874048/]
- 72. Comparative analysis, diversification, and functional ... [https://www.nature.com/articles/s41598-024-62876-5]
- 73. Comparative Evaluation of Computational Methods for ... [https://www.mdpi.com/2227-9059/13/8/2036]
- 74. Evaluation of candidate RT-qPCR reference genes in the ... [https://www.sciencedirect.com/science/article/pii/S2589958925000179]
- 75. Comparative analysis of RT-qPCR, flow cytometry, and Di ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12418842/]
- 76. Comparison of the Phenotypic Performance, Molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9824520/]
- 77. Efficient Virus-Induced Gene Silencing (VIGS) Method for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12115190/]
- 78. Establishment of a Virus-Induced Gene-Silencing (VIGS) ... [https://www.mdpi.com/1422-0067/24/1/392]
- 79. Virus-Induced Gene Silencing (VIGS): A Powerful Tool for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10057534/]
- 80. Development of a robust and efficient virus-induced gene ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01320-1]
- 81. Virus-induced gene silencing in plants [https://www.sciencedirect.com/science/article/abs/pii/S1046202303000379]
- 82. CRISPR Genome Engineering: Advantages and Limitations [https://www.taconic.com/resources/crispr-genome-engineering-advantages-limitations]
- 83. Applications of CRISPR/Cas tools in improving stress ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1616526/full]
- 84. Establishment of a CRISPR/Cas9 gene editing system based ... [https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-025-07094-5]
- 85. Comparative genomics‐driven design of virus‐delivered ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12576428/]
- 86. VIGE: virus-induced genome editing for improving abiotic ... [https://link.springer.com/article/10.1007/s44154-021-00026-x]
- 87. Development of Virus-Induced Gene Silencing (VIGS) and ... [https://dineshkumar.ucdavis.edu/research/development-virus-induced-gene-silencing-vigs-and-crisprcas9-mediated-genome-editing-approaches]
- 88. Prediction of plant complex traits via integration of multi- ... [https://www.nature.com/articles/s41467-024-50701-6]
- 89. Multi-omics Data Integration, Interpretation, and Its Application [https://pmc.ncbi.nlm.nih.gov/articles/PMC7003173/]
- 90. Spatial integration of multi-omics single-cell data with SIMO [https://www.nature.com/articles/s41467-025-56523-4]
- 91. Multi-omics network-based functional annotation of ... [https://pubmed.ncbi.nlm.nih.gov/34562334/]
- 92. Choosing the Right Tool for the Job: RNAi, TALEN or ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4441801/]
- 93. Gene Silencing – RNAi or CRISPR? [https://nordicbiosite.com/blog/gene-silencing-rnai-or-crispr]
- 94. VIGE: virus-induced genome editing for improving abiotic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10441944/]
- 95. A comparative analysis of high-throughput platforms for ... [https://www.nature.com/articles/srep10375]
- 96. Application and Expansion of Virus-Induced Gene ... [https://www.mdpi.com/2311-7524/9/8/934]