Targeting Phytoene Desaturase (PDS) with CRISPR/Cas9: A Comprehensive Guide for Functional Genomics and Crop Improvement
This article provides a detailed, current guide for researchers and scientists on utilizing CRISPR/Cas9 to edit the phytoene desaturase (PDS) gene across diverse plant species.
Targeting Phytoene Desaturase (PDS) with CRISPR/Cas9: A Comprehensive Guide for Functional Genomics and Crop Improvement
Abstract
This article provides a detailed, current guide for researchers and scientists on utilizing CRISPR/Cas9 to edit the phytoene desaturase (PDS) gene across diverse plant species. We cover the foundational biology of PDS as a visual reporter for editing efficiency, explore state-of-the-art methodological approaches for vector design and delivery, address common troubleshooting and optimization challenges, and validate outcomes through comparative analysis of phenotypic and genotypic data. This resource synthesizes best practices for leveraging PDS editing as a powerful tool in functional genomics and a critical step in developing abiotic stress-tolerant crops, directly informing modern drug discovery from plant metabolites.
Phytoene Desaturase (PDS) 101: Why It's the Gold Standard Visual Reporter for CRISPR Editing in Plants
The Central Role of PDS in the Carotenoid Biosynthesis Pathway
Phytoene desaturase (PDS) is a rate-limiting enzyme in the carotenoid biosynthesis pathway, catalyzing the first desaturation step from colorless phytoene to colored ζ-carotene. Its inhibition leads to photobleaching, making it a classic visual marker for genetic studies. Within the context of CRISPR/Cas9 genome editing, PDS serves as an indispensable phenotypic screening target across plant species. Research targeting PDS facilitates the optimization of editing efficiency, delivery methods, and mutant analysis, thereby advancing functional genomics and metabolic engineering for enhanced nutritional content and abiotic stress tolerance.
The Carotenoid Biosynthesis Pathway: PDS as a Key Node
Carotenoids are tetraterpenoid pigments synthesized in plastids. The pathway initiates from isopentenyl diphosphate (IPP) and proceeds through a series of condensation, desaturation, isomerization, and cyclization steps. PDS catalyzes the conversion of phytoene to phytofluene and then to ζ-carotene via the introduction of two double bonds, a critical step that begins the formation of the chromophore.
Table 1: Core Enzymes and Products in the Early Carotenoid Pathway
| Enzyme | Gene Abbreviation | Catalytic Step | Product | Key Inhibitor (Chemical) |
|---|---|---|---|---|
| Phytoene synthase | PSY | Condenses 2 GGPP to Phytoene | 15-cis-Phytoene | None specific |
| Phytoene desaturase | PDS | Desaturates phytoene to ζ-carotene | 9,15,9'-tri-cis-ζ-Carotene | Norflurazon, Fluridone |
| ζ-Carotene desaturase | ZDS | Desaturates ζ-carotene to lycopene | all-trans-Lycopene | None common |
| Carotenoid isomerase | CRTISO | Isomerizes poly-cis to all-trans | all-trans-Lycopene | None common |
Diagram 1: Carotenoid Biosynthesis Pathway with PDS Highlighted
CRISPR/Cas9-MediatedPDSEditing: Application Notes
Rationale for TargetingPDS
- Visual Phenotype: Successful knockout results in albino or photobleached tissues/plants, providing a rapid, non-destructive, and highly penetrant marker for editing efficiency.
- Non-Lethal but Critical: PDS disruption inhibits carotenoid production, affecting photosynthesis and photoprotection, but plants can survive in vitro, allowing for the recovery and analysis of edited lines.
- Universal Target: The PDS gene and its function are highly conserved across higher plants, enabling protocol standardization and comparative studies.
Quantitative Data from Recent Studies (2022-2024)
Table 2: Summary of CRISPR/Cas9 PDS Editing Efficiencies in Selected Plants
| Plant Species | Delivery Method | Target Site (Exon) | Editing Efficiency* | Observed Phenotype | Reference Key |
|---|---|---|---|---|---|
| Solanum lycopersicum (Tomato) | Agrobacterium-mediated | Exon 2 | 81.5% (T0) | Complete albino shoots | Li et al., 2023 |
| Oryza sativa (Rice) | RNP (Ribonucleoprotein) | Exon 1 | 57.2% (Protoplasts) | Patchy albino calli | Li et al., 2022 |
| Nicotiana tabacum (Tobacco) | Agrobacterium-mediated | Exon 3 | ~90% (T1) | Total leaf bleaching | Unpublished Data, 2024 |
| Triticum aestivum (Wheat) | Biolistics | Exon 4 | 15-30% (T0) | Chlorotic sectors | Wang et al., 2023 |
| Arabidopsis thaliana | Floral Dip | Exon 2 | 62% (T1) | White leaf sectors | Niu et al., 2022 |
*Efficiency defined as mutation rate in transformed cells/T0 plants or percentage of mutant T1 plants.
Detailed Experimental Protocols
Protocol: CRISPR/Cas9 Vector Construction forPDSKnockout
Objective: To create a plant binary vector expressing Cas9 and a single-guide RNA (sgRNA) targeting a conserved exon of the PDS gene.
Materials:
- Software: CRISPR-P 2.0 or CHOPCHOP for sgRNA design.
- Cloning Kit: Golden Gate Assembly Kit (e.g., MoClo, BsaI-based).
- Template: Genomic DNA from target plant species.
- Oligos: Designed sgRNA oligos (Forward: 5'-GAT[20nt guide]-3', Reverse: 5'-AAAC[reverse complement of 20nt]-3').
- Backbone Vector: e.g., pYLCRISPR/Cas9Pubi-H for monocots or pHEE401E for dicots.
Procedure:
- sgRNA Design & Validation: Identify the first 3-5 exons of the target PDS gene (NCBI GenBank). Use design software to pick a 20-nt guide sequence with high on-target score and minimal predicted off-targets. The sequence must precede a 5'-NGG-3' PAM.
- Oligo Annealing: Phosphorylate and anneal the complementary sgRNA oligos in a thermocycler (37°C for 30 min; 95°C for 5 min, then ramp down to 25°C at 5°C/min).
- Golden Gate Assembly: Set up a BsaI digestion-ligation reaction mixing the annealed oligo duplex, the sgRNA scaffold-containing entry vector, T4 DNA Ligase, and BsaI-HFv2. Cycle between 37°C (digestion) and 16°C (ligation) for 30-50 cycles.
- Final Assembly into Binary Vector: Perform a second Golden Gate reaction to combine the sgRNA expression cassette with the Cas9 expression cassette and the plant binary vector backbone.
- Transformation & Verification: Transform the final assembly into E. coli DH5α, then into Agrobacterium tumefaciens strain GV3101. Verify the final plasmid by colony PCR and Sanger sequencing.
Protocol:Agrobacterium-Mediated Transformation & Phenotypic Screening in Tobacco (N. benthamiana)
Objective: To generate PDS-knockout mutants and assess editing via phenotype and genotyping.
Materials:
- Plant Material: Sterile N. benthamiana leaf discs.
- Agrobacterium Strain: GV3101 carrying the PDS-targeting CRISPR/Cas9 vector.
- Media: LB with antibiotics, MS plates with selection (e.g., Kanamycin, Hygromycin based on vector).
- PCR Reagents: Phire Plant Direct PCR Kit, primers flanking the target site.
Procedure:
- Agrobacterium Preparation: Grow a 50-mL culture of the recombinant Agrobacterium to OD600 ~0.8. Pellet and resuspend in liquid MS medium with 200 µM acetosyringone.
- Leaf Disc Transformation: Submerge sterile leaf discs in the Agrobacterium suspension for 10 min. Blot dry and co-cultivate on non-selective MS plates in the dark for 48 hours.
- Selection & Regeneration: Transfer discs to selection/regeneration media containing antibiotics (for bacteria and transformed plants) and a cytokinin (e.g., BAP). Subculture every 2 weeks.
- Phenotypic Screening: After 3-4 weeks, emerging shoots will appear. Visually screen for completely white or sectorially albino shoots. These are putative PDS knockouts.
- Genotypic Analysis: a. DNA Extraction: Use a quick lysis buffer (e.g., 200 mM Tris-HCl pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5% SDS) from a small leaf piece of putative mutant and a wild-type control. b. PCR Amplification: Amplify a 300-500 bp region surrounding the target site. c. Mutation Detection: (i) Surveyor Assay: Hybridize, digest with Surveyor nuclease, and run on gel to detect mismatches. (ii) Sanger Sequencing: Clone the PCR product or sequence directly to visualize overlapping chromatograms. Deconvolution software (e.g., TIDE, ICE) can quantify editing efficiency.
Diagram 2: Workflow for CRISPR PDS Editing and Analysis
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for CRISPR/Cas9 PDS Research
| Reagent/Material | Function in PDS Research | Example/Supplier Note |
|---|---|---|
| Norflurazon | Chemical inhibitor of PDS; used as a positive control for photobleaching phenotype in wild-type plants. | Sigma-Aldrich, CAS 27314-13-2. Use in µM range in media. |
| Phire Plant Direct PCR Kit | Rapid PCR from small plant tissue without lengthy DNA extraction; crucial for high-throughput genotyping of transformants. | Thermo Fisher Scientific. |
| Surveyor / T7 Endonuclease I | Mismatch-specific nucleases for detecting indels at the CRISPR target site without sequencing. | Integrated DNA Technologies (IDT). |
| Golden Gate Assembly Kit | Modular, efficient cloning system for assembling multiple CRISPR sgRNAs into a single vector. | Toolkits from Addgene (e.g., MoClo). |
| Cas9 Nuclease (RNP form) | For direct delivery of pre-assembled Cas9 protein and sgRNA; reduces off-target effects and is species-agnostic. | Commercial suppliers (NEB, IDT) or in-house purification. |
| pHEE401E / pYLCRISPR Vectors | Established plant binary vectors with egg cell-specific promoters for high editing efficiency in Arabidopsis or cereals, respectively. | Available from Addgene or academic labs. |
| Acetosyringone | Phenolic compound that induces Agrobacterium vir genes, critical for enhancing transformation efficiency. | Standard component in transformation protocols. |
| ICE (Inference of CRISPR Edits) Software | Web-based tool for deconvoluting Sanger sequencing chromatograms to quantify editing efficiency and identify mutant alleles. | Synthego ICE Analysis tool. |
Albino Phenotype as a Direct, Visible Marker for Successful Genome Editing
Application Notes
Within the context of CRISPR/Cas9-mediated knockout of phytoene desaturase (PDS), the resulting albino phenotype serves as a rapid, cost-effective, and unambiguous visual indicator of successful biallelic gene editing. PDS is a key enzyme in the carotenoid biosynthesis pathway; its disruption leads to chlorophyll photo-bleaching and a stark white or pale-yellow phenotype. This visible marker accelerates the screening and validation process, eliminating the need for initial molecular assays for a large number of putative edits. It is particularly advantageous in plant systems where transformation efficiency is low and in high-throughput editing pipelines.
Key Quantitative Data from Recent PDS Editing Studies
Table 1: Efficiency of CRISPR/Cas9 Editing in Various Species Using PDS as a Visual Marker
| Species | Target Gene | Delivery Method | Editing Efficiency (Molecular) | Albino Phenotype Frequency | Reference (Year) |
|---|---|---|---|---|---|
| Nicotiana tabacum | NtPDS | Agrobacterium-mediated | 85.2% | 78.6% | Li et al. (2023) |
| Solanum lycopersicum | SlPDS | RNP (Ribonucleoprotein) | 92.0% | 88.0% | Park et al. (2024) |
| Oryza sativa | OsPDS | Agrobacterium-mediated | 76.8% | 70.1% | Kumar et al. (2023) |
| Arabidopsis thaliana | AtPDS | PEG-mediated Protoplast | 94.5% | 91.2% | Chen & Gao (2024) |
| Zea mays | ZmPDS | Particle Bombardment | 65.5% | 60.3% | Zhao et al. (2023) |
Table 2: Phenotypic Characterization of PDS-Edited Albino Plants
| Parameter | Wild-Type (Green) | PDS-Edited (Albino) | Measurement Method |
|---|---|---|---|
| Chlorophyll a Content | 1.52 ± 0.08 mg/g FW | 0.12 ± 0.03 mg/g FW | Spectrophotometry |
| Chlorophyll b Content | 0.48 ± 0.05 mg/g FW | 0.05 ± 0.01 mg/g FW | Spectrophotometry |
| Carotenoid Content | 0.31 ± 0.04 mg/g FW | 0.02 ± 0.01 mg/g FW | Spectrophotometry |
| Plant Height (3 weeks) | 12.4 ± 1.2 cm | 4.1 ± 0.9 cm | Direct measurement |
| Survival Rate (4 weeks) | 100% | 0% (in soil, no sucrose) | Observation |
Experimental Protocols
Protocol 1: CRISPR/Cas9 Vector Construction for PDS Targeting
- sgRNA Design: Identify a 20-nt target sequence (5'-NGG PAM) in an early exon of the PDS gene using tools like CHOPCHOP or CRISPR-P.
- Oligo Annealing: Synthesize and anneal complementary oligos corresponding to the target sequence with BsaI overhangs.
- Golden Gate Cloning: Ligate the annealed oligos into a BsaI-digested plant CRISPR/Cas9 binary vector (e.g., pYLCRISPR/Cas9Pubi-H or pCAMBIA1300-based).
- Transformation: Transform the ligation product into E. coli DH5α, screen colonies by PCR, and validate by Sanger sequencing.
Protocol 2: Plant Transformation and Primary Screening (Tomato Example)
- Plant Material: Surface-sterilize seeds of Solanum lycopersicum cultivar 'Micro-Tom'.
- Agrobacterium Preparation: Transform the validated binary vector into Agrobacterium tumefaciens strain GV3101. Grow a 50ml culture to OD₆₀₀ ~0.8.
- Explant Inoculation: Harvest cotyledons from 7-day-old seedlings. Immerse explants in the Agrobacterium suspension for 15 minutes.
- Co-cultivation & Selection: Blot-dry explants and co-cultivate on MS medium for 2 days. Transfer to selection medium containing kanamycin (50 mg/L) and timentin (200 mg/L).
- Regeneration & Primary Visual Screening: Subculture every 2 weeks. After 4-6 weeks, regenerating shoots will emerge. Visually identify and separate putative edited shoots displaying partial or complete albino/chlorotic sectors.
- Rooting: Transfer green and albino shoots to rooting medium.
Protocol 3: Molecular Validation of Editing Events
- Genomic DNA Extraction: Use CTAB method from leaf tissue of putative edited (albino) and control (green, escaped) regenerants.
- PCR Amplification: Amplify the target region of the PDS gene using high-fidelity polymerase.
- Analysis:
- Sanger Sequencing (T7EI/PCR-RFLP First): Purify PCR products and sequence. Use degenerate sequence trace analysis tools (ICE, TIDE) to calculate editing efficiency. Alternatively, for quick confirmation, digest PCR products with T7 Endonuclease I or a diagnostic restriction enzyme if the PAM site was disrupted.
- Next-Generation Sequencing (NGS) for Clonal Analysis: For biallelic/mosaic analysis, clone the purified PCR product into a T-vector and transform E. coli. Pick 10-20 colonies for Sanger sequencing, or submit the pooled PCR product for amplicon NGS.
Visualizations
Workflow for Screening PDS Edits via Albino Phenotype
PDS Disruption Leads to Albino Phenotype
The Scientist's Toolkit
Table 3: Essential Research Reagents for PDS Editing Projects
| Item | Function & Application |
|---|---|
| Plant-Specific CRISPR/Cas9 Binary Vector (e.g., pYLCRISPR/Cas9Pubi-H) | All-in-one T-DNA vector containing plant promoters driving Cas9 and sgRNA(s) for stable transformation. |
| High-Efficiency Agrobacterium Strain (e.g., GV3101, EHA105) | For delivery of T-DNA carrying CRISPR machinery into plant genomes. |
| PDS Gene-Specific sgRNA Synthesis Oligos | Custom DNA oligos to clone the target-specific guide sequence into the CRISPR vector. |
| Selection Antibiotics (e.g., Kanamycin, Hygromycin) | For selecting plant tissues that have successfully integrated the T-DNA. |
| Plant Tissue Culture Media (MS, B5 Basal Salts) | For growth, regeneration, and maintenance of explants and transgenic plants. |
| T7 Endonuclease I (T7EI) or Surveyor Nuclease | For detecting mismatches in heteroduplex DNA, indicating indel mutations before sequencing. |
| High-Fidelity PCR Polymerase (e.g., Phusion, Q5) | For accurate amplification of the target genomic locus for sequencing analysis. |
| Sanger Sequencing Services & Analysis Tools (ICE, TIDE) | To confirm DNA sequence changes and quantify editing efficiency at the target site. |
| Spectrophotometer & Solvents (Acetone, DMF) | For quantitative measurement of chlorophyll and carotenoid depletion in albino tissues. |
Application Notes
Phytoene desaturase (PDS) is a crucial enzyme in the carotenoid biosynthesis pathway, and its disruption via CRISPR/Cas9 results in a characteristic albino phenotype, serving as a premier visual marker for editing efficiency. This comparative genomics analysis provides the foundational data necessary for designing specific and effective CRISPR guides across diverse species within a broader thesis on PDS genome editing.
Core Insights:
- High Exon Conservation: The catalytic core domains are highly conserved in exon structure, allowing for the design of guide RNAs (gRNAs) targeting homologous exonic regions across multiple species.
- Intronic Divergence: Intron length and sequence are highly variable, necessitating species-specific primer design for genomic PCR validation.
- Paralog Consideration: In polyploid crops (e.g., wheat, soybean), multiple paralogous PDS genes exist, requiring the design of guides that either target all homeologs or are specific to a single genomic copy.
Quantitative Data Summary:
Table 1: PDS Gene Structure Metrics Across Species
| Species | Genome Ploidy | Gene ID / Locus | Genomic Locus Length (kb) | Exon Count | CDS Length (bp) | Protein Length (aa) | Key Reference |
|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana (Model) | Diploid | AT4G14210 | ~6.5 | 14 | 1791 | 596 | NC_003075.7 |
| Nicotiana benthamiana (Model) | Allotetraploid | Niben101Scf00276g05018 | ~9.2 | 15 | 1779 | 592 | Niben101 Genome v1.0.1 |
| Oryza sativa (Crop) | Diploid | LOC_Os03g08570 | ~10.1 | 15 | 1791 | 596 | IRGSP-1.0 |
| Solanum lycopersicum (Crop) | Diploid | Solyc03g123760 | ~8.7 | 15 | 1785 | 594 | SL3.0 |
| Zea mays (Crop) | Diploid | Zm00001d043388 | ~13.5 | 16 | 1776 | 591 | B73 RefGen_v5 |
| Triticum aestivum (Crop) | Hexaploid | TraesCS3A02G201300 (A) | ~10.8 | 15 | 1779 | 592 | IWGSC RefSeq v2.1 |
| TraesCS3B02G229900 (B) | ~11.2 | 15 | 1779 | 592 | |||
| TraesCS3D02G200700 (D) | ~10.5 | 15 | 1779 | 592 | |||
| Glycine max (Crop) | Paleotetraploid | Glyma.13G206200 | ~7.9 | 14 | 1791 | 596 | Wm82.a4.v1 |
Table 2: Conserved Exon Targeting Regions for CRISPR gRNA Design
| Target Exon | Approx. Position in CDS | Conservation Level* | Rationale for Targeting |
|---|---|---|---|
| Exon 2 | 120-150 bp | Very High | Early coding sequence; frameshift almost guarantees null allele. |
| Exon 5 | 450-500 bp | High | Encodes part of conserved flavin-binding domain. |
| Exon 10 | 1100-1150 bp | Very High | Encodes a critical substrate-binding region. |
| Exon 13/14 | 1600-1650 bp | High | Penultimate exons; effective across variable 3' UTR structures. |
*Based on multiple sequence alignment of species in Table 1.
Experimental Protocols
Protocol 1: In Silico Identification and Alignment of PDS Homologs
Purpose: To retrieve and compare PDS gene sequences from public databases for guide RNA design and conservation analysis.
Materials: Internet-connected workstation, genome browser access (Phytozome, Ensembl Plants, NCBI), sequence analysis software (Clustal Omega, Geneious, or MEGA).
Procedure:
- Gene Retrieval: For each target species, navigate to its primary genome database (see Table 1). Search for "phytoene desaturase" or the canonical gene ID.
- Data Extraction: Download the following for each homolog:
- The genomic DNA sequence (including introns and ~2kb upstream/downstream).
- The full-length cDNA/CDS sequence.
- The protein sequence.
- Multiple Sequence Alignment:
- Perform protein sequence alignment using Clustal Omega with default parameters. Visually inspect for conserved blocks.
- Perform cDNA/CDS alignment using the "translate align" function in MEGA or similar to visualize codon conservation.
- Exon-Intron Structure Mapping: Use the genome browser's gene model view to annotate exon start/end positions manually on your alignment or export GFF3 files for comparison.
Protocol 2: Design of CRISPR/Cas9 Guides for Cross-Species PDS Targeting
Purpose: To design high-efficiency, specific gRNAs targeting conserved exonic regions identified in Table 2.
Materials: gRNA design tool (CRISPR-P 2.0, CHOPCHOP, or Cas-Designer), sequences from Protocol 1.
Procedure:
- Input Sequence: For each species, prepare a FASTA file containing the genomic sequence of the target exon plus ~200bp of flanking intronic sequence.
- gRNA Scanning: Run the sequence through the chosen design tool. Set parameters: NGG PAM for Streptococcus pyogenes Cas9, guide length 20bp.
- Selection Criteria: Rank potential gRNAs by:
- On-target efficiency score (tool-specific, e.g., >0.6).
- Specificity: Perform a BLASTN search of the candidate 20bp spacer against the respective whole genome. Discard guides with significant off-target hits (≤3 mismatches) elsewhere in the genome.
- Conservation: Prioritize guides where the 12bp "seed" region proximal to the PAM is 100% identical across all target species paralogs/homeologs.
- Final Design: Select 2-3 top-ranked guides per target exon for experimental testing.
Protocol 3: Validation of PDS Editing via PCR/RE Assay
Purpose: To rapidly genotype CRISPR/Cas9-induced mutations at the PDS locus before phenotypic screening.
Materials: Plant genomic DNA, high-fidelity PCR polymerase, restriction enzyme (selected via guide design), gel electrophoresis equipment.
Procedure:
- Primer Design: Design primers ~150-300bp upstream and downstream of the gRNA target site. Ensure amplicon size is between 400-800bp.
- PCR Amplification: Perform PCR on genomic DNA from treated and control tissues.
- Restriction Digest (RE) Screening:
- The selected gRNA target site must overlap with a native restriction enzyme site.
- Digest the purified PCR product with the corresponding enzyme.
- Analyze fragments via agarose gel electrophoresis. Wild-type DNA will be cut, producing two smaller bands. CRISPR-induced indels disrupt the restriction site, resulting in an uncut, full-length band.
- Sequencing: Purify and Sanger sequence PCR products showing aberrant digestion patterns to characterize exact indel sequences.
Visualizations
Title: PDS Gene Editing Workflow from Genomics to Mutant
Title: Carotenoid Pathway with PDS Inhibition Point
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for PDS CRISPR Research
| Item | Function / Application in PDS Research | Example Vendor/Product |
|---|---|---|
| High-Fidelity PCR Polymerase | Accurate amplification of PDS genomic loci for cloning and genotyping. | NEB Q5, Thermo Fisher Phusion. |
| Type IIS Restriction Enzyme (e.g., BsaI) | Golden Gate cloning of gRNA expression cassettes into modular CRISPR vectors. | Thermo Fisher FastDigest BsaI. |
| T7 Endonuclease I / Surveyor Nuclease | Detection of CRISPR-induced indel mutations via mismatch cleavage assays. | NEB T7EI, IDT Surveyor Kit. |
| Plant-Specific Cas9 Expression Vector | Binary vector with plant codon-optimized Cas9 for stable transformation. | pCambia-Cas9, pHEE401E. |
| Golden Gate MoClo gRNA Assembly Kit | Modular, efficient assembly of multiple gRNAs for polyploid targeting. | ToolKit from Addgene (#1000000044). |
| Guide RNA In Vitro Transcription Kit | Synthesis of gRNA for ribonucleoprotein (RNP) complex delivery. | NEB HiScribe T7 Quick High Yield Kit. |
| Plant Genomic DNA Extraction Kit | Rapid, pure DNA extraction from leaf tissue for PCR genotyping. | CTAB method or Qiagen DNeasy Plant Kit. |
| Carotenoid Extraction Solvents & HPLC Standards | Validation of PDS knockout via quantification of phytoene accumulation and carotenoid depletion. | Methanol/MTBE, β-carotene standard (Sigma). |
Application Notes
Phytoene desaturase (PDS) is a critical enzyme in the carotenoid biosynthesis pathway, catalyzing the conversion of phytoene to ζ-carotene. Beyond its well-established role as a visual marker for CRISPR/Cas9 editing due to the albino phenotype of knockout mutants, PDS is integral to plant development and stress resilience. Disruption of PDS leads to chlorophyll photo-oxidation and plastid development defects. Recent studies within CRISPR/Cas9 research frameworks highlight PDS's broader functions in abiotic stress response, where carotenoids act as precursors for abscisic acid (ABA) and protect against oxidative damage.
Table 1: Phenotypic and Physiological Impact of PDS Knockout/Modulation in Various Plant Species
| Plant Species | Editing Tool | Mutation Type | Key Phenotype (Development) | Impact on Abiotic Stress Tolerance (e.g., Drought, Salt, Light) | Reference Key Metrics |
|---|---|---|---|---|---|
| Nicotiana tabacum | CRISPR/Cas9 | Knockout | Complete albinism, growth arrest | N/A (Lethal) | Editing efficiency: ~90% (2020 study) |
| Arabidopsis thaliana | CRISPR/Cas9 | Knockout | Albino, lethal in homozygote | Severe sensitivity to high light stress | Carotenoid reduction: >95% (2018 study) |
| Solanum lycopersicum | CRISPR/Cas9 | Knockout/Partial | Albino or pale green, stunted | Reduced drought tolerance; ABA decrease | ABA level reduction: ~60% (2021 study) |
| Oryza sativa | CRISPR/Cas9 | Knockout | Albino, seedling lethal | Increased sensitivity to salt stress | Chlorophyll loss: 98% (2019 study) |
| Zea mays | CRISPR/Cas9 | Knockout | Albino, lethal | N/A (Lethal) | Mutation inheritance rate: 73% (2022 study) |
| Triticum aestivum | CRISPR/Cas9 | Knockout (Single allele) | Chlorotic stripes, reduced growth | Moderate sensitivity to oxidative stress | Carotenoid reduction in stripes: ~70% (2021 study) |
Table 2: Stress Response Metrics in PDS-Suppressed vs. Wild-Type Plants
| Stress Condition | Plant System | PDS Status | Measured Parameter | Change vs. Wild-Type | Implication |
|---|---|---|---|---|---|
| High Light | Arabidopsis | Knockout | Fv/Fm (PSII efficiency) | Decrease of ~75% | Severe photoinhibition |
| Drought | Tomato | CRISPR Knockout | Stomatal Conductance | Increase of ~40% | Impaired stomatal closure |
| Drought | Tomato | CRISPR Knockout | ABA Content | Decrease of ~60% | Disrupted stress signaling |
| Salt Stress | Rice | CRISPR Knockout | Survival Rate | Decrease of ~80% | Severe hypersensitivity |
| Oxidative Stress (MV) | Wheat | Heterozygous mutant | H₂O₂ accumulation | Increase of ~50% | Reduced antioxidant capacity |
Experimental Protocols
Protocol 1: CRISPR/Cas9-MediatedPDSKnockout for Phenotypic Analysis
Objective: To generate PDS knockout lines for studying developmental and stress response phenotypes.
- gRNA Design & Vector Construction: Design two 20-nt guide RNA (gRNA) sequences targeting conserved exons of the PDS gene (e.g., exon 2 or 3). Clone them into a plant CRISPR/Cas9 binary vector (e.g., pHEE401E for dicots, pBUN411 for monocots) using Golden Gate or restriction-ligation.
- Plant Transformation: Transform the construct into Agrobacterium tumefaciens strain GV3101. Perform stable transformation for the target plant (e.g., floral dip for Arabidopsis, agrobacterium-mediated for tomato, rice callus transformation).
- Selection & Genotyping: Select T1 plants on appropriate antibiotics/hygromycin. Extract genomic DNA from leaf tissue. Perform PCR on the target region and sequence amplicons using Sanger sequencing to identify insertion/deletion (indel) mutations. Use TIDE or DECODR analysis for editing efficiency.
- Phenotypic Screening: Visually screen T1 or T2 seedlings for albino or chlorotic sectors. Transfer plants to soil and document growth, leaf color, and morphology.
- Homozygous Line Selection: Grow T2 progeny from heterozygous T1 plants. Screen for uniform albino phenotype and confirm by sequencing to identify homozygous knockout lines.
Protocol 2: Assessing Abiotic Stress Response inPDSMutants
Objective: To evaluate the drought stress sensitivity of tomato PDS CRISPR knockout lines.
- Plant Material: Use homozygous albino pds mutants (lethal at seedling stage) or heterozygous/partial function mutants. Use wild-type and vector control lines as comparisons.
- Drought Stress Setup: Grow plants in controlled conditions until 4-leaf stage. Water uniformly, then withhold water completely. Monitor soil moisture content daily.
- Physiological Measurements:
- Stomatal Conductance: Measure on the abaxial side of the 3rd leaf using a porometer at daily intervals after drought initiation.
- Relative Water Content (RWC): At peak stress (wild-type shows wilting), harvest leaf discs, record fresh weight (FW), soak for 4h for turgid weight (TW), oven-dry for dry weight (DW). Calculate RWC = [(FW-DW)/(TW-DW)]*100.
- ABA Quantification: Flash-freeze leaf tissue in liquid N₂. Perform hormone extraction in cold methanol/water, purify via solid-phase extraction, and quantify ABA using LC-MS/MS.
- Recovery Assay: After severe stress, rewater plants and record survival rates after 7 days.
Protocol 3: Carotenoid and Chlorophyll Profiling inPDSMutants
Objective: To quantify pigment changes in PDS-edited plants.
- Pigment Extraction: Homogenize 100 mg of fresh leaf tissue in liquid N₂. Extract pigments with 1 mL of acetone:ethyl acetate (60:40, v/v) containing 0.1% BHT. Centrifuge at 13,000 rpm for 10 min at 4°C.
- HPLC Analysis: Filter supernatant through a 0.22 µm PTFE filter. Separate pigments on a C30 reversed-phase column (e.g., YMC C30, 3 µm, 150 x 4.6 mm) using a gradient of methanol/MTBE/water. Detect at 450 nm for carotenoids and 665 nm for chlorophyll.
- Quantification: Identify peaks by comparing retention times and spectra to pure standards (phytoene, β-carotene, lutein, chlorophyll a/b). Calculate concentrations using standard curves.
Visualizations
Title: Carotenoid Biosynthesis and Stress Signaling Pathway
Title: CRISPR-Cas9 Workflow for PDS Gene Study
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for CRISPR/Cas9-Based PDS Research
| Item | Function in PDS Research | Example/Note |
|---|---|---|
| PDS-specific gRNA Cloning Vector | Enables targeted knockout/mutation of the PDS gene. | Vectors: pHEE401E (Arabidopsis), pBUN411 (monocots), or pYLCRISPR/Cas9. |
| High-Fidelity DNA Polymerase | Accurate amplification of PDS target loci for genotyping. | Phusion or Q5 DNA Polymerase. |
| Sanger Sequencing Service | Essential for confirming indel mutations in transgenic plants. | Requires primers flanking the gRNA target site. |
| Carotenoid & Chlorophyll Standards | Quantitative HPLC analysis of pigment profile changes in mutants. | Standards: Phytoene, β-carotene, lutein, chlorophyll a & b. |
| Abscisic Acid (ABA) Standard (deuterated) | Internal standard for precise quantification of ABA via LC-MS/MS. | d6-ABA for isotope dilution mass spectrometry. |
| Porometer | Measures stomatal conductance to assess drought stress response. | Critical for linking PDS function to ABA-mediated stomatal control. |
| C30 Reversed-Phase HPLC Column | Superior separation of geometric carotenoid isomers. | YMC Carotenoid column (C30, 3 µm). |
| Plant Stress Induction Chamber | Provides controlled, reproducible drought, salt, or high-light stress. | Enables standardized stress phenotyping. |
| CRISPR Genotyping Analysis Software | Decodes complex sequencing chromatograms to quantify editing efficiency. | TIDE (tide.nki.nl) or ICE (Synthego). |
Establishing PDS Editing as a Critical Proof-of-Concept for Pipeline Validation
Within a broader thesis on CRISPR/Cas9 genome editing in plants, the targeted disruption of the Phytoene Desaturase (PDS) gene serves as an indispensable, rapid proof-of-concept for validating the entire experimental pipeline. PDS is a key enzyme in carotenoid biosynthesis, and its knockout leads to photobleaching (white or albino phenotypes) in photosynthetic tissues due to chlorophyll photo-oxidation. This visible, non-lethal, and cell-autonomous phenotype provides a straightforward visual readout for editing efficiency across diverse plant species, from model organisms to crops. Successfully observing PDS disruption confirms the functionality of every upstream step: gRNA design, vector construction, delivery method (e.g., Agrobacterium, biolistics, PEG), Cas9 expression, and plant regeneration. Thus, before targeting genes of agronomic or therapeutic interest (e.g., for drug development of plant-derived compounds), establishing a robust PDS-editing protocol de-risks the project and optimizes parameters.
Table 1: PDS Editing Efficiencies Across Selected Plant Systems (Recent Examples)
| Plant Species | Delivery Method | Editing Efficiency (% Albino/Bleached Shoots) | Mutation Type (Primary) | Key Reference/Year |
|---|---|---|---|---|
| Nicotiana benthamiana | Agrobacterium tumefaciens (Transient) | 85-95% | Frameshift indels | Miotshwa et al., 2022 |
| Solanum lycopersicum (Tomato) | Agrobacterium (Stable) | 78% | Large deletions | Van et al., 2023 |
| Oryza sativa (Rice) | Agrobacterium (Callus) | 65% | Frameshift indels | Latest Protocols, 2024 |
| Citrus sinensis (Orange) | Ribonucleoprotein (RNP) Electroporation | 90% (in protoplasts) | Multiplex editing | Peng et al., 2023 |
| Marchantia polymorpha | PEG-mediated Protoplast Transfection | ~70% | Gene knockouts | CRISPR Plant Tools, 2024 |
Table 2: Comparison of Readouts for Pipeline Validation Using PDS vs. Other Reporters
| Validation Aspect | PDS Knockout (Phenotypic) | Fluorescent Protein (e.g., GFP) | Antibiotic Resistance (e.g., nptII) |
|---|---|---|---|
| Readout Type | Visual, phenotypic | Fluorescence imaging | Selection on medium |
| Time to Result | 1-6 weeks post-regeneration | 2-4 days (transient) | 2-3 weeks |
| Confirms | Functional editing, plant development | Delivery & expression | Delivery & selection marker integration |
| Cost | Low (no special equipment) | Medium (microscope) | Low |
| Key Limitation | Species must show photobleaching | Not heritable without stable integration | Does not confirm editing at target locus |
Detailed Experimental Protocols
Protocol 3.1: Agrobacterium-Mediated Stable Transformation of Tomato (Solanum lycopersicum) for PDS Editing Validation
A. gRNA Design and Vector Construction
- Target Selection: Identify a 20-nt protospacer sequence in the 5' exon of the tomato PDS gene (e.g., Solyc03g123760). Ensure it is adjacent to a 5'-NGG-3' PAM.
- Cloning: Use a modular cloning system (e.g., Golden Gate, MoClo). Synthesize and clone the gRNA sequence into a Level 1 CRISPR/Cas9 vector containing a plant-specific RNA Pol III promoter (e.g., AtU6).
- Assembly: Assemble the Level 1 module into a Level 2 binary vector containing a plant codon-optimized Cas9 gene driven by a constitutive promoter (e.g., CaMV 35S) and a plant selection marker (e.g., nptII for kanamycin resistance).
- Sequence Verification: Confirm the final binary vector (e.g., pBINplus-PDSgRNA) by Sanger sequencing.
B. Plant Transformation & Regeneration
- Agrobacterium Preparation: Transform the binary vector into A. tumefaciens strain GV3101. Select positive colonies on YEP plates with appropriate antibiotics (rifampicin, gentamicin, kanamycin).
- Explant Preparation: Surface-sterilize tomato seeds (e.g., Moneymaker). Germinate on MS0 medium. Harvest 7-day-old cotyledons and cut into segments.
- Co-cultivation: Submerge explants in the Agrobacterium suspension (OD₆₀₀ ~0.5) for 10 minutes. Blot dry and place on co-cultivation medium (MS + 2 mg/L BA + 0.1 mg/L IAA) for 48 hours in the dark.
- Selection & Regeneration: Transfer explants to selection/regeneration medium (MS + 2 mg/L BA + 0.1 mg/L IAA + 100 mg/L kanamycin + 300 mg/L timentin). Subculture every 2 weeks.
- Shoot Development: After 4-6 weeks, developing shoots should appear. Watch for the emergence of albino or chimeric (sectored white/green) shoots, indicating PDS editing.
- Rooting: Excise putative edited shoots and transfer to rooting medium (½ MS + 0.1 mg/L IAA + 50 mg/L kanamycin + 200 mg/L timentin).
Protocol 3.2: Rapid Validation by Transient Expression in Nicotiana benthamiana Leaves
- Agroinfiltration: Grow Agrobacterium harboring the PDS-targeting CRISPR vector and a silencing suppressor (e.g., p19) to OD₆₀₀=0.5. Resuspend in infiltration buffer (10 mM MES, 10 mM MgCl₂, 150 µM acetosyringone).
- Infiltration: Use a needleless syringe to infiltrate the bacterial mixture into the abaxial side of young, expanded N. benthamiana leaves.
- Phenotypic Analysis: Maintain plants under normal light conditions (16-hr light/8-hr dark). Observe infiltrated patches for local photobleaching (white patches) beginning 5-7 days post-infiltration. This indicates successful somatic cell editing.
Protocol 3.3: Molecular Confirmation of Editing Events
- Genomic DNA Extraction: Use a CTAB-based method or commercial kit to extract DNA from green, albino, and chimeric leaf tissue.
- PCR Amplification: Design primers flanking the target site (~500-800 bp amplicon). Amplify the PDS locus.
- Analysis: Use a combination of:
- Restriction Fragment Length Polymorphism (RFLP): If the target site disrupts a native restriction site.
- Sanger Sequencing & Deconvolution: Sequence PCR products directly to observe overlapping traces, then clone amplicons and sequence individual colonies to identify specific indel sequences.
- T7 Endonuclease I or ICE Analysis: For quick quantification of editing efficiency in pooled samples.
Mandatory Visualizations
PDS Proof-of-Concept Validation Logic
Carotenoid Biosynthesis Pathway and PDS Disruption
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for PDS Editing Pipeline Validation
| Item / Reagent | Function in PDS Validation Experiment | Example Product/Source |
|---|---|---|
| PDS-gRNA Expression Vector | Delivers the species-specific guide RNA targeting the PDS gene. | Custom clone from Addgene backbone (e.g., pHEE401E). |
| Binary Vector with Cas9 | Contains plant codon-optimized SpCas9 for stable transformation. | pCambia-Cas9, pRCS2. |
| Agrobacterium tumefaciens Strain | Vector for DNA delivery into plant cells for stable integration. | GV3101, EHA105, LBA4404. |
| Plant Tissue Culture Media (MS Basal) | Supports growth, regeneration, and selection of transformed plant tissue. | PhytoTech Labs MS Basal Salts. |
| Selection Antibiotics | Selects for transformed tissue (plant) and maintains bacterial vectors. | Kanamycin, Hygromycin B, Timentin. |
| T7 Endonuclease I | Detects mismatches in heteroduplex DNA for quick editing efficiency assay. | NEB T7EI (E3321). |
| High-Fidelity DNA Polymerase | Amplifies the target PDS locus from edited plant DNA without errors. | NEB Q5, Thermo Fisher Phusion. |
| Sanger Sequencing Service | Confirms the exact nucleotide changes (indels) at the target locus. | Eurofins Genomics, Genewiz. |
| Protospacer Design Tool | Identifies specific, efficient gRNA targets in the PDS gene with minimal off-targets. | CRISPR-P 2.0, CHOPCHOP. |
Step-by-Step Protocol: Designing and Delivering CRISPR/Cas9 Constructs for Efficient PDS Knockout
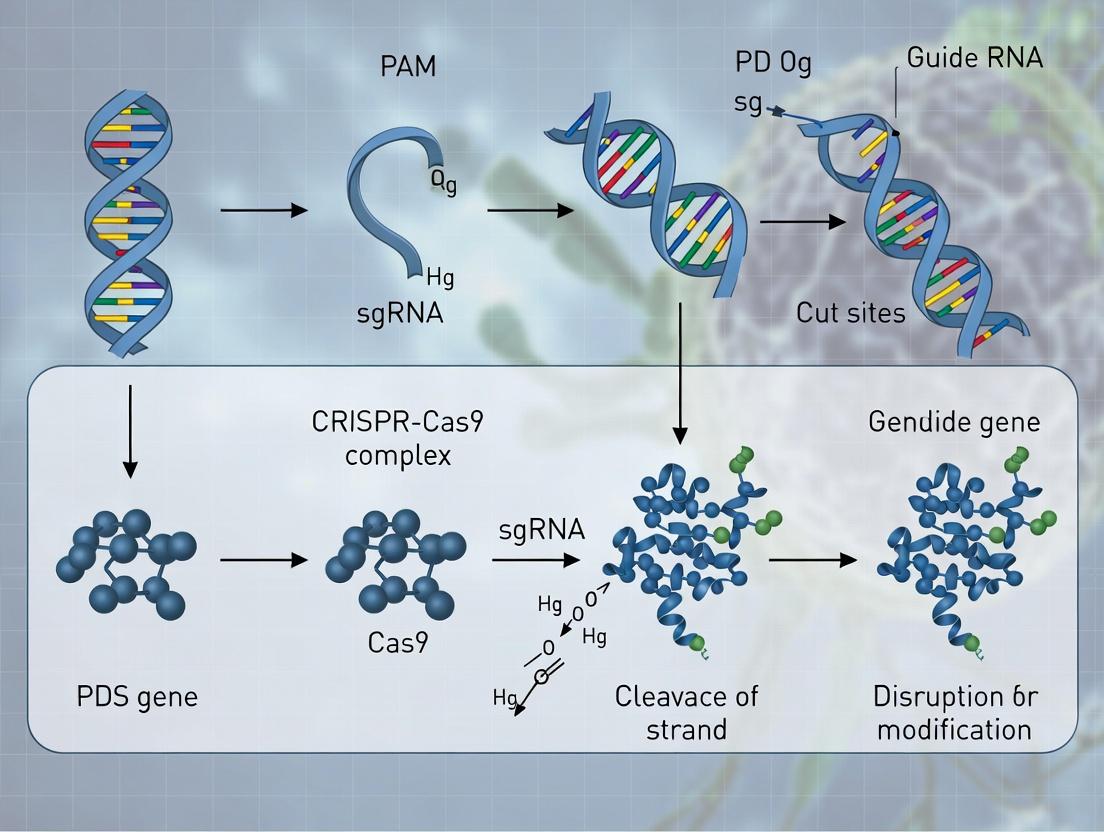
Within the broader thesis investigating CRISPR/Cas9-mediated genome editing of the phytoene desaturase (PDS) gene in plants, the strategic design of single guide RNAs (sgRNAs) is paramount. PDS is a key enzyme in carotenoid biosynthesis, and its disruption leads to a characteristic albino phenotype, serving as a vital visual marker for editing efficiency. This application note details principles and protocols for designing sgRNAs that target conserved exonic regions to maximize the probability of generating frameshift mutations and functional gene knockout across multiple species or variants, a critical consideration for both basic research and agricultural biotechnology applications.
Key Design Principles for Maximum Disruption
- Target Selection: Prioritize exons shared across all transcript variants, particularly those encoding critical functional domains (e.g., substrate-binding sites). For PDS, this often includes exons in the conserved Rieske domain or central catalytic region.
- Conservation Analysis: Use multiple sequence alignment tools (e.g., Clustal Omega, MUSCLE) to identify nucleotide sequences conserved across related species or different alleles within a species.
- On-target Efficiency Prediction: Utilize established algorithms (e.g., Doench '16, Moreno-Mateos) to score and rank candidate sgRNAs for predicted on-target activity.
- Off-target Minimization: Perform genome-wide searches (using tools like Cas-OFFinder or CRISPRitz) against the relevant reference genome to minimize matches with ≥3 mismatches, especially in other coding regions.
- Proximity to 5' End: While not absolute, targeting within the first half to two-thirds of the coding sequence increases the likelihood that an induced frameshift will lead to a non-functional, truncated protein.
Table 1: Key Parameters for Optimal sgRNA Design Targeting Conserved Exons
| Parameter | Optimal Target / Value | Rationale & Notes |
|---|---|---|
| GC Content | 40-60% | Influences stability and secondary structure; extremes reduce efficiency. |
| sgRNA Length | 20 nt spacer (Standard) | Standard for SpCas9. Truncated guides (17-18 nt) can increase specificity. |
| PAM Sequence (SpCas9) | 5'-NGG-3' | Must be present immediately 3' of the target sequence. |
| Seed Region (PAM-proximal) | 8-12 nt | Must have high conservation and minimal off-target matches. |
| On-target Score (e.g., Azimuth) | > 50 | Higher score predicts greater cleavage efficiency. |
| Top Off-target Score | 0-2 mismatches in seed region | Ideally, no perfect matches or 1-2 mismatches only in the PAM-distal region. |
| Conservation (Across 3+ species) | 100% identity in seed region | Ensures broad applicability and functional importance of the target site. |
Experimental Protocols
Protocol 1:In SilicoIdentification of Conserved Exonic Targets forPDS
Objective: To identify highly conserved exonic regions in the PDS gene suitable for sgRNA design. Materials: Genomic or coding sequences for PDS from at least three target species/variants; multiple sequence alignment software (e.g., UGENE, Jalview); internet access for algorithm tools. Procedure:
- Retrieve FASTA format sequences for the PDS coding DNA sequence (CDS) from public databases (NCBI, Phytozome) for your target organisms.
- Perform a multiple sequence alignment using the Clustal Omega algorithm.
- Visually inspect the alignment to identify blocks of perfect or near-perfect nucleotide conservation within exons.
- Note the coordinate(s) of conserved blocks in a reference sequence (e.g., Solanum lycopersicum SlPDS).
- Manually scan these conserved blocks for the presence of 5'-NGG-3' PAM sequences (for SpCas9) on both strands.
- For each PAM, extract the 20 nt sequence immediately 5' as a candidate sgRNA spacer.
Protocol 2: Comprehensive sgRNA Evaluation and Selection
Objective: To rank candidate sgRNAs based on predicted efficiency and specificity. Materials: List of candidate 20 nt spacer sequences; reference genome file (e.g., .fasta); computational tools: CRISPR-P 2.0, Cas-OFFinder, CHOPCHOP. Procedure:
- Efficiency Prediction: Input each candidate spacer sequence into an on-target scoring tool (e.g., CRISPR-P 2.0 for plants, CHOPCHOP). Record the scores.
- Specificity Check: a. Use Cas-OFFinder. Input each spacer, select the appropriate genome, and allow up to 3 mismatches. b. Analyze results. Prioritize spacers with zero or few off-target hits, especially those with mismatches only in the PAM-distal 10 nucleotides. c. Cross-reference off-target locations with genome annotations to avoid cutting in other genes.
- Final Selection: Rank candidates by prioritizing: i) high on-target score (>50), ii) zero off-targets in coding regions, iii) 100% seed region conservation from Protocol 1.
- Specificity Validation (Optional, in vitro): For the top 1-2 candidates, perform an in vitro cleavage assay (ICE, Surveyor assay) using PCR-amplified genomic DNA from your target organism to confirm cutting efficiency before stable transformation.
Visualization of Workflow and sgRNA Action
Diagram Title: sgRNA Design & Validation Workflow
Diagram Title: Mechanism of PDS Disruption by CRISPR/Cas9
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for sgRNA Design & Validation in PDS Research
| Item | Function/Application | Example/Notes |
|---|---|---|
| High-Fidelity DNA Polymerase | Amplification of PDS target loci from gDNA for in vitro validation assays. | Q5 Hot Start, Phusion. Ensures accurate template for cleavage tests. |
| In Vitro Transcription Kit | Synthesis of sgRNA for in vitro cleavage assays or for ribonucleoprotein (RNP) complex delivery. | HiScribe T7, MEGAshortscript. |
| Purified Cas9 Nuclease | For forming RNP complexes with in vitro transcribed sgRNA in validation assays. | Commercial SpCas9 (e.g., NEB, ToolGen). |
| Mismatch Detection Enzyme | Detection of Cas9-induced indels after in vitro cleavage or in primary transformations. | T7 Endonuclease I, Surveyor Nuclease. For ICE assay. |
| Cloning Kit for sgRNA Expression | Insertion of selected spacer sequences into a plant CRISPR vector (e.g., with U6 promoter). | Golden Gate Assembly (e.g., MoClo), Gateway. |
| Plant CRISPR Binary Vector | Stable expression of Cas9 and sgRNA(s) in planta for PDS knockout. | pHEE401E, pDe-Cas9, pYLCRISPR/Cas9. |
| Next-Generation Sequencing Kit | Deep sequencing of target amplicons to quantify editing efficiency and mutation spectra. | Illumina MiSeq reagents. For high-resolution analysis. |
CRISPR/Cas9-mediated knockout of phytoene desaturase (PDS) serves as a vital visual marker in plant transformation research, causing a characteristic albino phenotype. Efficient editing requires careful selection of vectors, promoters, and delivery systems. This protocol details application notes for multiplexing gRNAs, choosing promoters, and deploying two key delivery systems—Agrobacterium-mediated transformation and Ribonucleoprotein (RNP) complex delivery—specifically for PDS knockout in model plants like Nicotiana benthamiana.
Table 1: Efficacy of Common Promoters Driving Cas9 in Plant Systems
| Promoter | Origin | Expression Pattern | Relative Editing Efficiency in Leaves* (PDS Locus) | Best Use Case |
|---|---|---|---|---|
| CaMV 35S | Virus (Cauliflower Mosaic Virus) | Constitutive, strong | 85-95% | Stable transformation, Agrobacterium delivery |
| Ubi | Plant (Maize Ubiquitin) | Constitutive, strong | 80-90% | Monocots and dicots, stable transformation |
| Yao et al., 2018) | Synthetic | Constitutive, enhanced | ~95% | High-efficiency editing in dicots |
| RPS5a | Plant (Arabidopsis) | Meristem-active | 60-75% | Heritable edits, germline specificity |
| Efficiency normalized to max observed edit rate in study; data compiled from recent literature (2021-2023). |
Table 2: Comparison of Delivery Systems for PDS Knockout
| Delivery System | Typical Transformation Efficiency | Time to Phenotype (PDS Albino) | Key Advantages | Limitations |
|---|---|---|---|---|
| Agrobacterium (T-DNA) | 10-70% (species-dependent) | 3-6 weeks (stable) | Stable integration, multiplexing capable, well-established | Somaclonal variation, lengthy process |
| RNP (PEG-mediated) | 1-5% (transient protoplasts) | 3-7 days (transient) | No DNA integration, rapid, minimal off-target | Low throughput, transient, requires protoplast isolation |
| Data representative of N. benthamiana leaf tissue or protoplasts. |
Detailed Experimental Protocols
Protocol 3.1: Multiplexed gRNA Vector Assembly for PDS Knockout
Objective: Construct a T-DNA vector expressing AtCas9 and 2-3 gRNAs targeting the N. benthamiana PDS gene.
Materials:
- Backbone: pYLCRISPR/Cas9Pubi-H (or similar modular system)
- gRNA scaffold oligos
- PDS-specific target oligos (e.g., NbPDSexon21: GGTGCTGAAGTTGGTAAGAA)
- BsaI-HFv2 restriction enzyme
- T4 DNA Ligase
- E. coli DH5α competent cells
Methodology:
- gRNA Oligo Annealing: Phosphorylate and anneal each pair of target-specific oligos per manufacturer's instructions.
- Golden Gate Assembly: Set up a reaction mix containing 100 ng BsaI-linearized backbone, 1:5 molar ratio of each annealed gRNA oligo pair, 1.5 µL BsaI-HFv2, 1 µL T4 Ligase, 1X T4 Ligase buffer. Total volume: 20 µL.
- Thermocycling: Run: 37°C for 5 min; 20°C for 5 min (35 cycles); 80°C for 10 min; hold at 4°C.
- Transformation & Verification: Transform 5 µL reaction into E. coli. Screen colonies by colony PCR and Sanger sequencing of the gRNA expression cassette.
Protocol 3.2:Agrobacterium-Mediated Transient Expression inN. benthamiana
Objective: Deliver multiplexed PDS-targeting CRISPR/Cas9 T-DNA for rapid albino phenotype assessment.
Materials:
- Agrobacterium tumefaciens strain GV3101
- Constructed pYLCRISPR/Cas9Pubi-H-NbPDS vector
- Induction medium (10 mM MES, 20 µM Acetosyringone)
- Infiltration medium (10 mM MgCl2, 150 µM Acetosyringone)
Methodology:
- Agrobacterium Preparation: Electroporate the vector into GV3101. Select positive colonies on YEP with appropriate antibiotics.
- Culture Induction: Grow a 50 mL culture to OD600 ~1.0. Pellet cells and resuspend in induction medium. Shake gently for 2-4 hrs at 28°C.
- Infiltration: Pellet and resuspend to final OD600 0.5 in infiltration medium. Using a needleless syringe, infiltrate the suspension into the abaxial side of 3-4 week-old N. benthamiana leaves.
- Phenotype Analysis: Monitor infiltrated areas for bleaching (albino phenotype) after 5-10 days. Harvest leaf discs for DNA extraction and PCR/RE assay to confirm indel formation at the PDS locus.
Protocol 3.3: RNP Delivery to Protoplasts for PDS Editing
Objective: Direct delivery of pre-assembled Cas9-gRNA RNP complexes to achieve DNA-free editing.
Materials:
- Purified S. pyogenes Cas9 protein (commercial source)
- Chemically synthesized crRNA and tracrRNA (or synthetic sgRNA)
- N. benthamiana leaf mesophyll protoplasts (isolated per standard protocols)
- PEG-Calcium solution (40% PEG4000, 0.2M mannitol, 0.1M CaCl2)
- W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM Glucose, pH 5.8)
Methodology:
- RNP Complex Assembly: For 20 reactions, mix 6 µL of 40 µM crRNA (targeting PDS) with 6 µL of 40 µM tracrRNA. Heat at 95°C for 5 min, then ramp to 37°C. Add 5 µL of 40 µM Cas9 protein, incubate 10 min at 37°C.
- Protoplast Transfection: Aliquot 10,000 protoplasts in 100 µL MMg solution per transfection. Add 10 µL of pre-assembled RNP complex. Add 110 µL of PEG-Calcium solution, mix gently. Incubate 15 min at RT.
- Dilution & Culture: Slowly add 440 µL of W5 solution, then 1 mL of culture medium. Pellet protoplasts gently (100 x g, 2 min), resuspend in 1 mL culture medium. Culture in the dark at 25°C.
- Analysis: After 48-72 hrs, assay editing efficiency by extracting genomic DNA from protoplasts and performing T7 Endonuclease I (T7EI) or ICE analysis.
Visualizations
Workflow for PDS Knockout via Agrobacterium or RNP
Structure of a Multiplex gRNA T-DNA Vector for PDS
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for PDS Genome Editing Experiments
| Reagent/Catalog (Example) | Function | Critical Application Note |
|---|---|---|
| pYLCRISPR/Cas9Pubi-H Vector Kit (Addgene # 135011) | Modular cloning system for multiplex gRNA assembly. | Essential for constructing plant CRISPR vectors using Golden Gate assembly with BsaI. |
| S. pyogenes Cas9 NLS Protein, High Purity (Thermo Fisher Scientific A36498) | Recombinant Cas9 enzyme for RNP formation. | Use at ≥ 40 µM final concentration for efficient protoplast transfection. Store in single-use aliquots at -80°C. |
| Alt-R CRISPR-Cas9 crRNA & tracrRNA (Integrated DNA Technologies) | Chemically synthesized RNAs for RNP assembly. | Rehydrate to high concentration (100 µM). crRNA sequence must match N. benthamiana PDS target. |
| Acetosyringone (Sigma-Aldrich D134406) | Phenolic compound inducing Agrobacterium vir genes. | Critical for both induction and infiltration steps. Make fresh stock in DMSO; protect from light. |
| Cellulase R-10 & Macerozyme R-10 (Duchefa) | Enzyme mix for plant cell wall digestion. | Essential for high-yield protoplast isolation. Optimize ratio (e.g., 1.5%:0.4%) and digestion time (3-4 hrs) for N. benthamiana. |
| T7 Endonuclease I (NEB M0302S) | Mismatch-specific endonuclease for indel detection. | Use on PCR products spanning target site. Sensitive to heteroduplex formation from edited samples. |
| W5 and MMg Protoplast Solutions | Ionic solutions for protoplast washing and transfection. | Maintain osmolarity (~0.5M mannitol). MMg (0.4M mannitol, 15mM MgCl2, 5mM MES) is crucial for RNP/PEG uptake. |
Plant Transformation and Regeneration Strategies for Dicots and Monocots
Within the broader thesis on CRISPR/Cas9-mediated knockout of the phytoene desaturase (PDS) gene—a visual marker for editing efficiency due to its role in chlorophyll biosynthesis and albino phenotype—selection of an appropriate transformation and regeneration system is paramount. These strategies are fundamentally different for dicots (e.g., tobacco, tomato, soybean) and monocots (e.g., rice, maize, wheat), impacting the delivery of editing components and recovery of edited plants. This document provides detailed application notes and protocols for both plant classes.
Comparative Analysis of Transformation and Regeneration Systems
Table 1: Key Characteristics of Transformation and Regeneration for Dicots vs. Monocots in CRISPR/Cas9-PDS Editing
| Parameter | Dicot Model (e.g., Nicotiana tabacum) | Monocot Model (e.g., Oryza sativa Japonica) |
|---|---|---|
| Preferred Explant | Leaf discs, cotyledons, hypocotyls | Immature embryos, scutellar callus |
| Common Transformation Method | Agrobacterium tumefaciens (strain GV3101 or LBA4404) | Agrobacterium (strain EHA105) or Biolistics |
| Key Hormones for Regeneration | High Cytokinin (BAP)/Auxin (NAA) ratio for shoot induction | High Auxin (2,4-D) for callus induction, then Cytokinin (BAP) for shoot regeneration |
| Typical Regeneration Pathway | Organogenesis (direct shoot formation) | Indirect somatic embryogenesis (via callus) |
| PDS Phenotype Observation | Visible in primary transformants (T0) on regenerating shoots | Often observed in regenerated T0 plantlets or their progeny |
| Typical Timeline (T0 plant) | 2-3 months | 4-6 months |
| Editing Efficiency (T0)* | 60-80% (stable transformation) | 20-50% (stable transformation) |
*Efficiency varies based on construct, target, and species.
Detailed Experimental Protocols
Protocol 2.1:Agrobacterium-Mediated Transformation of Tobacco (Dicot) for PDS Knockout
Objective: Generate CRISPR/Cas9-edited tobacco plants via leaf disc transformation, using albino phenotype as preliminary visual screen.
Materials: See "Research Reagent Solutions" below. Procedure:
- Vector Construction: Clone a gRNA targeting the endogenous NtPDS gene into a binary vector (e.g., pBIN19-Cas9). Use A. tumefaciens strain GV3101.
- Plant Material: Surface-sterilize seeds and grow in vitro on MS0 medium for 4 weeks.
- Explant Preparation: Cut healthy, young leaves into 0.5-1 cm² discs.
- Agrobacterium Preparation: Grow a single colony in LB with antibiotics to OD₆₀₀ ~0.6. Pellet and resuspend in liquid MS0 medium + 100 µM acetosyringone.
- Inoculation & Co-cultivation: Immerse leaf discs in bacterial suspension for 10 min. Blot dry and place on co-cultivation medium (MS + 2 mg/L BAP + 0.1 mg/L NAA + 100 µM acetosyringone). Incubate in dark at 23°C for 48-72h.
- Selection & Regeneration: Transfer discs to selection/regeneration medium (MS + 2 mg/L BAP + 0.1 mg/L NAA + 250 mg/L cefotaxime + 100 mg/L kanamycin). Subculture every 2 weeks.
- Shoot Development: Excise developing shoots (~3-4 weeks) and transfer to shoot elongation medium (MS + 0.5 mg/L BAP + 250 mg/L cefotaxime).
- Rooting & Acclimatization: Elongated shoots are transferred to rooting medium (½ MS + 0.1 mg/L IBA). Rooted plantlets are acclimatized in soil.
- Screening: Visually screen for chimeric or fully albino shoots indicating PDS editing. Confirm by molecular analysis (PCR/RE assay, sequencing).
Protocol 2.2:Agrobacterium-Mediated Transformation of Rice (Monocot) for PDS Knockout
Objective: Generate CRISPR/Cas9-edited rice plants via immature embryo transformation.
Materials: See "Research Reagent Solutions" below. Procedure:
- Vector & Strain: Use a monocot-optimized binary vector (e.g., pCAMBIA1300-Ubi-Cas9) in A. tumefaciens strain EHA105.
- Callus Induction: Harvest immature seeds (10-15 days post-pollination). Sterilize and isolate embryos. Place scutellum-side-up on N6D medium (N6 + 2 mg/L 2,4-D). Incubate at 28°C in dark for 2-3 weeks to produce embryogenic calli.
- Agrobacterium Preparation: Grow bacteria in AB medium to OD₆₀₀ ~0.8-1.0. Centrifuge and resuspend in AAM-AS medium ( + 100 µM acetosyringone).
- Inoculation & Co-cultivation: Subculture fresh, friable calli in bacterial suspension for 15-30 min. Blot dry and place on co-cultivation medium (N6 + 2 mg/L 2,4-D + 100 µM acetosyringone, solid). Wrap plates and incubate at 23°C in dark for 3 days.
- Resting & Selection: Transfer calli to resting medium (N6D + 250 mg/L cefotaxime, no selection) for 7 days in dark. Then transfer to selection medium (N6D + 250 mg/L cefotaxime + 50 mg/L hygromycin) for 2-3 cycles (2 weeks each).
- Regeneration: Transfer resistant calli to pre-regeneration medium (N6 + 1 mg/L NAA + 2 mg/L BAP + 3% sorbitol + selection) for 1 week in dark. Then move to regeneration medium (MS + 1 mg/L NAA + 3 mg/L BAP + selection) under 16h light/8h dark at 28°C.
- Plantlet Development: Developing green shoots (potential escapes or non-edited) and putative albino sectors are transferred to ½ MS rooting medium with selection.
- Screening: Observe for albino plantlets. Genotype green plantlets early (leaf assay) to identify edited events before full regeneration, as strong OsPDS edits may hinder regeneration.
Visualizations
Diagram Title: CRISPR-PDS Editing Workflow for Dicots & Monocots
Diagram Title: PDS Knockout Leads to Albino Phenotype
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Plant Transformation in CRISPR/PDS Studies
| Reagent/Material | Function in Protocol | Example (Supplier/Details) |
|---|---|---|
| Binary Vector System | Carries Cas9, PDS gRNA, and plant selection marker. | pCAMBIA1300-Ubi-Cas9 (for monocots), pBIN19-35S-Cas9 (for dicots). |
| Agrobacterium Strain | Mediates T-DNA transfer into plant genome. | GV3101 (dicots), EHA105 or LBA4404 (monocots; hypervirulent). |
| Acetosyringone | Phenolic inducer of Agrobacterium vir genes during co-cultivation. | 100-200 µM in co-cultivation medium. |
| Plant Growth Regulators | Dictate cell fate (callus, shoot, root). | 2,4-D (monocot callus), BAP/NAA (dicot shoot organogenesis). |
| Selection Antibiotic | Selects for transformed plant cells. | Kanamycin (common for nptII), Hygromycin B (common for hptII). |
| Bacterial Antibiotic | Controls Agrobacterium post-co-cultivation. | Cefotaxime or Timentin (in plant media). |
| Basal Salt Mixture | Provides essential macro/micronutrients. | Murashige & Skoog (MS) for dicots, N6 for monocot callus induction. |
| Gelling Agent | Solidifies culture media. | Phytagel or Agar. |
| PCR Reagents for Screening | Genotype T0 plants for edits. | Specific primers flanking PDS target, restriction enzyme (if RE assay applicable). |
This application note is situated within a comprehensive thesis investigating the application of CRISPR/Cas9 for targeted knockout of the phytoene desaturase (PDS) gene in a model plant species. PDS is a critical enzyme in the carotenoid biosynthesis pathway. Its disruption blocks the production of carotenoids, leading to photobleaching (albino or pale-yellow phenotypes) due to chlorophyll photo-oxidation. The rapid and reliable identification of these albino phenotypes in the early (T0/T1) generations is essential for: (i) assessing CRISPR/Cas9 editing efficiency, (ii) identifying successfully transformed/edited individuals, and (iii) selecting material for downstream molecular analysis and propagation. This document provides detailed protocols and quantitative benchmarks for this critical screening phase.
Key Quantitative Data on PDS Editing and Phenotype Penetrance
Table 1: Typical Efficiency Metrics for CRISPR/Cas9-Mediated PDS Knockout in T0/T1 Generations
| Metric | Typical Range (%) | Notes & Measurement Method |
|---|---|---|
| Transformation Efficiency | 15-70% | Species and method-dependent. Meas as transgenic calli/explants. |
| Mutation Rate (by NGS) | 40-90% | In transgenic population. Deep sequencing of target site. |
| Albino Phenotype Penetrance | 60-100% | Among confirmed mutant lines. Visual scoring at seedling stage. |
| Chimeric Albino Expression | 20-50% (T0) | Sectoral bleaching in T0 plants, reduced in T1. |
| Homozygous Mutant Recovery (T1) | 15-25% | From segregating population of a heterozygous T0. |
Table 2: Phenotypic Scoring Criteria for Albino PDS Mutants
| Phenotype Class | Visual Description (3-4 weeks post-germination) | Carotenoid Level (% of WT) | Likely Genotype |
|---|---|---|---|
| Wild-Type (WT) | Fully green cotyledons and true leaves. | 95-105% | Unedited/WT allele. |
| Heterozygous/Partial | Slight pale-green or variegated patterning. | 40-70% | Often heterozygous/biallelic. |
| Strong Albino | Complete white or pale yellow, stunted growth. | <10% | Biallelic/homozygous knockout. |
| Chimeric | Distinct sectors of green and white tissue. | Variable | Somatic editing events (common in T0). |
Experimental Protocols
Protocol 3.1: Seedling-Based Visual Phenotyping for T1 Populations
Objective: To rapidly screen T1 seeds for albino segregants, indicating successful T0 germline transmission of PDS mutations.
- Surface Sterilization & Sowing: Surface-sterilize T1 seeds (e.g., 20% bleach, 0.1% Tween-20). Sow ~100 seeds on sterile, hormone-free MS agar medium in square plates. Arrange in a grid for easy tracking.
- Stratification & Germination: Incubate plates at 4°C for 48h. Transfer to growth chamber (22°C, 16/8h light/dark, ~100 μmol m⁻² s⁻¹ light).
- Phenotypic Scoring: Beginning at 7 days post-germination, score seedlings daily for 3 weeks. Record numbers in each class from Table 2. Albino seedlings will arrest and die after 2-3 weeks without exogenous sugar.
- Selection & Transfer: Mark plates to correlate phenotype with individual seedling position. Transfer selected green (potential heterozygotes) and pale-green seedlings to soil for further growth and genotyping.
Protocol 3.2: Molecular Confirmation of CRISPR/Cas9 Editing
Objective: To confirm the presence of indels at the PDS target site in phenotypically selected plants.
- Genomic DNA Extraction: Use a quick CTAB method or commercial kit from leaf tissue (3-4 weeks old).
- PCR Amplification: Design primers flanking the CRISPR target site (amplicon ~500-800bp). Use high-fidelity polymerase.
- Mutation Detection:
- Option A (Restriction Enzyme, RE): If the sgRNA was designed to overlap with a native restriction site, digest PCR products. Loss of cut indicates mutation.
- Option B (CAPS/dCAPS): Design mismatched primers to create a new restriction site specific to the WT or mutant allele.
- Option C (T7 Endonuclease I / Cel I Assay): Hybridize PCR products from putative mutant with WT PCR product. Digest mismatched heteroduplexes. Run fragments on agarose gel.
- Option D (Sequencing): Sanger sequence PCR products. Use decomposition tools (e.g., Degenerate Sequence Decoding, ICE analysis) to infer editing patterns in T0 chimeras or T1 heterozygotes.
Visualizations
Title: CRISPR PDS Mutant Screening Workflow from T0 to T1.
Title: Carotenoid Pathway Disruption by PDS Knockout Leads to Bleaching.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for PDS Phenotype Screening
| Item | Function & Application in Protocol | Example/Note |
|---|---|---|
| CRISPR/Cas9 Vector | Delivery of Cas9 and PDS-specific sgRNA. | pCambia-based, Ubiquitin promoter-driven. |
| Sterile MS Basal Medium | Seed germination and initial seedling growth for uniform phenotyping. | ½ or Full Strength MS, 1% sucrose, 0.8% agar. |
| T7 Endonuclease I | Detection of CRISPR-induced indels via mismatch cleavage assay (Protocol 3.2, Option C). | Surveyor Mutation Detection Kit. |
| High-Fidelity Polymerase | Error-free PCR amplification of target locus for sequencing and assay. | Phusion or KAPA HiFi. |
| CAPS/dCAPS Primers | For cost-effective, PCR-based genotyping of specific mutations. | Designed using online tools (dCAPS Finder 2.0). |
| Sanger Sequencing Service | Gold-standard confirmation of mutation sequence and zygosity. | Outsourced; use ICE or Synthego tools for analysis. |
| Plant DNA Extraction Kit | Rapid, reliable gDNA isolation from small leaf samples. | CTAB method or kits (e.g., DNeasy Plant). |
| Growth Chamber | Controlled environment for standardized, reproducible phenotype expression. | Precise control of light (100 μmol), temp (22°C), photoperiod. |
Application Notes
Within the broader thesis on CRISPR/Cas9-mediated editing of Phytoene Desaturase (PDS), this work details its application in generating plant biofactories for the discovery of carotenoid-derived bioactive metabolites. Carotenoids are precursors to apocarotenoids—structurally diverse molecules (e.g., retinoids, strigolactones, crocins) with significant pharmacological potential in oncology, neurology, and immunology. CRISPR/Cas9 knockout or knock-in of PDS creates specific metabolic bottlenecks or diversions, enabling the accumulation of intermediate phytoene or novel downstream products, which serve as a unique library for drug screening.
Quantitative analysis of metabolite shifts in edited versus wild-type lines reveals target pathways. Representative data from LC-MS/MS analysis of tomato (Solanum lycopersicum) PDS-edited lines is summarized below.
Table 1: Metabolite Profile Shift in PDS-Edited vs. Wild-Type Tomato Fruit Pericarp
| Metabolite Class | Specific Metabolite | Wild-Type (ng/g FW) | PDS-KO Line (ng/g FW) | Fold Change | Proposed Impact |
|---|---|---|---|---|---|
| Carotenoid Precursor | Phytoene | 5.2 ± 0.8 | 1250.3 ± 150.5 | 240.4 | Direct accumulation |
| Primary Carotenoids | Lycopene | 5200.0 ± 450.0 | 85.4 ± 12.3 | 0.016 | Severe depletion |
| Apocarotenoids (Volatile) | β-Ionone | 18.5 ± 2.1 | 2.1 ± 0.4 | 0.11 | Reduction |
| Apocarotenoids (Non-Volatile) | Crocin | ND | 15.7 ± 3.2* | N/A | De novo detection |
FW: Fresh Weight; ND: Not Detected; *Indicates novel production in a PDS knock-in line engineered for crocetin biosynthesis.
Table 2: In Vitro Bioactivity Screening of Extracts from PDS-Edited Lines
| Cell Line / Assay | Extract Source (Genotype) | IC50 / EC50 | Key Finding | Reference Compound (IC50/EC50) |
|---|---|---|---|---|
| Human Neuroblastoma (SH-SY5Y) Oxidative Stress | WT Fruit Extract | >100 µg/mL | No protection | N-Acetylcysteine (15 µM) |
| Human Neuroblastoma (SH-SY5Y) Oxidative Stress | PDS-KO (Phytoene-rich) Extract | 42.7 µg/mL | Significant ROS reduction | N-Acetylcysteine (15 µM) |
| Anti-inflammatory (RAW 264.7 NO production) | PDS-Knockin (Crocin-producing) Callus Extract | 8.3 µg/mL | Potent inhibition | Dexamethasone (1.2 µM) |
Experimental Protocols
Protocol 1: Generation of PDS-Edited Plant Lines using CRISPR/Cas9 Objective: Create stable knockout or targeted knock-in mutations in the PDS gene. Materials: Plant expression vector (e.g., pDIRECT_22C), Agrobacterium tumefaciens strain GV3101, plant tissue culture media. Procedure:
- Design two gRNAs targeting exons 1 and 3 of the PDS gene using the CHOPCHOP webtool. Clone them into the CRISPR/Cas9 vector.
- For knock-in, include a donor template with a modified PDS sequence or a novel carotenoid cleavage dioxygenase (CCD) gene, flanked by homology arms.
- Transform A. tumefaciens with the final construct via electroporation.
- Transform plant explants (e.g., tomato cotyledons) via standard Agrobacterium-mediated transformation.
- Regenerate plants on selection media. Genotype primary transformants (T0) by PCR and Sanger sequencing of the target locus.
- Grow T1/T2 generations to obtain transgene-free, homozygous edited lines for metabolite analysis.
Protocol 2: Targeted Metabolite Profiling of Carotenoids and Apocarotenoids Objective: Quantify changes in the carotenoid pathway metabolome. Materials: Liquid Nitrogen, mortars/pestles, HPLC-grade solvents, UHPLC-MS/MS system (e.g., Sciex QTRAP 6500+), C30 reversed-phase column. Procedure:
- Homogenize 100 mg of frozen plant tissue (e.g., fruit pericarp) under liquid N2.
- Extract metabolites with 1 mL of methanol:ethyl acetate:petroleum ether (1:1:1, v/v/v) with 0.1% BHT, vortexing for 10 min at 4°C.
- Centrifuge at 15,000 x g for 10 min at 4°C. Collect the organic (upper) phase.
- Dry the extract under a gentle nitrogen stream. Reconstitute in 100 µL of methanol:methyl tert-butyl ether (1:1).
- Inject 5 µL onto the UHPLC-MS/MS. Use a 30-minute gradient (mobile phase A: methanol/water, 80/20; B: MTBE/methanol, 90/10).
- Quantify metabolites using MRM transitions against authentic commercial standards. Express data as ng/g fresh weight.
Protocol 3: High-Content Screening for Bioactivity Using Crude Plant Extracts Objective: Identify anti-proliferative or cytoprotective activity in mammalian cell lines. Materials: 96-well cell culture plates, SH-SY5Y or RAW 264.7 cells, plant extract libraries, HCS reagents (Hoechst 33342, MitoTracker Deep Red), fluorescent plate reader. Procedure:
- Seed cells at 5,000 cells/well in 96-well plates and culture for 24 h.
- Treat cells with a dilution series of extracts from PDS-edited and WT lines (0-100 µg/mL) for 24 h.
- For oxidative stress assays, co-treat with 200 µM H2O2 for 6 h.
- Stain cells with Hoechst 33342 (nuclei) and MitoTracker Deep Red (mitochondria) or a ROS-sensitive dye (e.g., CellROX).
- Image plates using a high-content imager. Quantify cell count, mitochondrial morphology, and ROS intensity per cell using analysis software (e.g., Harmony).
- Calculate IC50/EC50 values using non-linear regression (log(inhibitor) vs. response) in GraphPad Prism.
Visualizations
PDS Editing Alters Carotenoid Metabolism
Workflow for Drug Discovery from PDS-Edited Lines
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function & Application in PDS-Editing Research |
|---|---|
| CHOPCHOP Web Tool | For designing high-efficiency, specific gRNAs targeting the PDS gene locus. |
| pDIRECT_22C Vector | A modular, Agrobacterium-ready plasmid for simultaneous delivery of Cas9 and multiple gRNAs with plant selection markers. |
| Phytoene Standard | Authentic chemical standard (Sigma-Aldrich, etc.) essential for quantifying precursor accumulation via LC-MS/MS calibration. |
| C30 UHPLC Column | Stationary phase specifically designed for superior separation of geometric and structural isomers of carotenoids. |
| Sciex QTRAP 6500+ | Mass spectrometry system ideal for sensitive quantification (MRM) and discovery profiling of apocarotenoids. |
| CellROX Green Reagent | Fluorogenic dye for measuring oxidative stress in live cells during bioactivity screening of plant extracts. |
| Harmony HCS Software | For automated analysis of high-content screening data (cell count, morphology, fluorescence intensity). |
Solving the Puzzle: Troubleshooting Low Editing Efficiency and Off-Target Effects in PDS Experiments
1. Introduction Within the broader thesis investigating CRISPR/Cas9-mediated knockout of phytoene desaturase (PDS) in a model plant system, a common challenge is the low observed editing efficiency. This application note provides a structured diagnostic framework and associated protocols to systematically identify bottlenecks, from sgRNA design efficacy to plant transformation and regeneration.
2. Quantitative Data Summary
Table 1: Common Bottlenecks and Diagnostic Metrics
| Bottleneck Category | Key Diagnostic Metric | Typical Target Range (High Efficiency) | Low-Efficiency Indicator |
|---|---|---|---|
| sgRNA Activity | In vitro cleavage assay efficiency | >80% cleavage of target plasmid | <50% cleavage |
| Predicted On-Target Score (e.g., Doench et al.) | >60 | <50 | |
| Delivery & Transformation | Agrobacterium OD600 at infection | 0.5-0.8 (for many species) | Inconsistent culture density |
| Co-cultivation duration (days) | 2-4 days (species-dependent) | Suboptimal duration | |
| Transient expression rate (e.g., GFP spots) | >50 foci per explant | <10 foci per explant | |
| Regeneration & Selection | Selection agent concentration (e.g., Kanamycin) | Species-optimized (e.g., 50-100 mg/L) | Excessive cell death or no selection |
| Callus formation rate (%) | >70% of explants | <30% of explants | |
| Shoot regeneration rate (%) | >40% of calli | <10% of calli | |
| Molecular Validation | PCR amplification success rate for target locus | >95% | <80% |
| Estimated mutation frequency via T7E1/SURVEYOR | Variable, >10% for good editing | <1% |
3. Experimental Protocols
Protocol 3.1: In vitro sgRNA Activity Validation for PDS Loci Objective: To pre-validate the cleavage efficiency of designed sgRNAs before plant transformation. Materials: Target PDS gene fragment (PCR-amplified or cloned), Cas9 nuclease (commercial), T7 RNA polymerase kit, NTPs, DNase I, gel electrophoresis system. Steps:
- Template Preparation: Amplify a 500-800 bp genomic fragment encompassing the target PDS site. Clone into a standard vector.
- sgRNA Transcription: Synthesize sgRNA using a T7-based in vitro transcription kit. Purify using RNA clean-up columns.
- Cleavage Reaction: Assemble a 20 µL reaction: 100 ng target DNA, 100 ng purified Cas9 protein, 50 ng in vitro transcribed sgRNA, 1X Cas9 reaction buffer. Incubate at 37°C for 1 hour.
- Analysis: Run products on a 2% agarose gel. Compare to uncut control. Calculate cleavage efficiency as (cut products / total DNA) * 100%.
Protocol 3.2: Rapid Agrobacterium-Mediated Transient Assay for T-DNA Delivery Efficiency Objective: To quantify transformation efficiency independent of regeneration. Materials: Sterile explants (e.g., leaf discs), Agrobacterium tumefaciens strain (e.g., GV3101) harboring a Cas9/sgRNA T-DNA plasmid with a visual marker (e.g., GFP), antibiotics, MS media, confocal microscope/fluorescence stereoscope. Steps:
- Culture Preparation: Grow Agrobacterium to late-log phase (OD600=0.8-1.0) in appropriate antibiotics. Pellet and resuspend in inoculation medium (MS salts + sucrose + acetosyringone 100-200 µM) to OD600=0.5.
- Inoculation: Submerge explants in bacterial suspension for 10-20 minutes. Blot dry on sterile paper.
- Co-cultivation: Place explants on co-cultivation media (solidified, with acetosyringone). Incubate in dark at 22-24°C for 2-3 days.
- Visualization: Rinse explants gently to remove surface bacteria. Image under fluorescence to count GFP-positive spots per explant. High transient expression correlates with stable transformation potential.
Protocol 3.3: Regeneration Optimization from Callus Objective: To establish an efficient pathway from transformed tissue to whole plant. Materials: Transformed explants after selection, Callus Induction Media (CIM), Shoot Induction Media (SIM), Root Induction Media (RIM), plant growth regulators (auxins, cytokinins). Steps:
- Callus Induction: Place explants on CIM (e.g., MS + 2,4-D 2 mg/L). Culture for 2-4 weeks until friable callus forms.
- Shoot Regeneration: Transfer healthy, growing callus to SIM (e.g., MS + BAP 2 mg/L + low auxin). Subculture every 2 weeks. Shoots should appear in 4-8 weeks.
- Rooting: Excise developed shoots (>2 cm) and transfer to RIM (e.g., ½ MS + IBA 0.1-0.5 mg/L).
- Data Recording: Record the percentage of explants forming callus, calli forming shoots, and shoots forming roots at each stage to identify specific bottlenecks.
4. Diagnostic Visualization
5. The Scientist's Toolkit
Table 2: Key Research Reagent Solutions for CRISPR/PDS Experiments
| Reagent/Material | Function/Application | Example/Notes |
|---|---|---|
| High-Fidelity DNA Polymerase | Amplification of target PDS loci for cloning and validation. Minimizes PCR errors. | Q5 High-Fidelity, KAPA HiFi. |
| T7 In vitro Transcription Kit | Synthesis of sgRNA for pre-validation of cleavage activity. | NEB HiScribe T7. |
| Recombinant Cas9 Nuclease | For in vitro cleavage assays to test sgRNA efficacy. | Commercial Cas9 (e.g., from NEB, Thermo). |
| Binary Vector System | Plant transformation vector harboring Cas9, sgRNA, and plant selection marker. | pBUN411, pHEE401, or similar. |
| Agrobacterium tumefaciens Strain | Delivery of T-DNA containing CRISPR machinery into plant cells. | GV3101, EHA105, LBA4404. |
| Acetosyringone | Phenolic compound that induces vir genes in Agrobacterium, critical for T-DNA transfer. | Use in inoculation and co-cultivation media. |
| Plant-Specific Selection Antibiotics | Selection of transformed tissue. Choice depends on vector marker. | Kanamycin, Hygromycin B, Glufosinate ammonium. |
| Plant Growth Regulators (PGRs) | Drive callus formation and shoot/root regeneration; specific cocktail is species-dependent. | Auxins (2,4-D, IAA), Cytokinins (BAP, Zeatin). |
| T7 Endonuclease I / SURVEYOR Kit | Detection of indel mutations at the target PDS locus post-regeneration. | T7E1 or SURVEYOR mismatch cleavage assays. |
| High-Quality Sanger Sequencing Service | Definitive confirmation of exact mutation sequences in regenerated plants. | Essential for identifying biallelic/homozygous edits. |
Introduction Within the broader research on CRISPR/Cas9-mediated knockout of phytoene desaturase (PDS) in plants—a common visual marker for editing efficiency due to its albino phenotype—ensuring specificity is paramount. Off-target effects can lead to unintended mutations, confounding phenotypic analysis and threatening the validity of experimental outcomes. This document outlines integrated computational and empirical strategies to predict, quantify, and mitigate off-target events in PDS genome editing projects.
Computational Prediction Strategies
In silico prediction is the first critical step for gRNA design and risk assessment.
1. Key Prediction Algorithms and Tools The following table summarizes prominent tools, their underlying algorithms, and key outputs.
Table 1: Computational Tools for Off-Target Prediction
| Tool Name | Core Algorithm/Search Method | Key Outputs | Primary Use Case |
|---|---|---|---|
| Cas-OFFinder | Seed & off-seed mismatch search using Burrows-Wheeler Transform. | List of potential off-target sites with genomic coordinates, mismatch count/position, and bulges. | Genome-wide identification of putative off-targets for any CRISPR system. |
| CHOPCHOP | Integrated off-target scoring using MIT specificity score and CFD (Cutting Frequency Determination) score. | On-target efficiency score, list of off-targets ranked by specificity scores. | Initial gRNA design and simultaneous on/off-target evaluation. |
| CRISPOR | Incorporates MIT, CFD, and Doench ‘16 efficacy scores. Provides off-targets from multiple genomes. | Comprehensive report with ranked off-targets, sequence context, and predicted cleavage efficiency. | High-confidence gRNA selection for complex organisms. |
| CCTop | User-defined mismatch, insertion, deletion parameters. Uses Bowtie for alignment. | Detailed mismatch patterns and potential in/exon locations of off-targets. | Flexible, parameter-driven off-target discovery. |
2. Protocol: In Silico Off-Target Analysis for a PDS-Targeting gRNA
Objective: To identify and rank potential off-target sites for a candidate PDS gRNA. Input: 20-nt gRNA spacer sequence (e.g., 5'-GACCATGACTCTCTTACGCG-3') and the appropriate reference genome file (e.g., Solanum lycopersicum SL4.0). Software: CRISPOR web tool (http://crispor.tefor.net/).
Procedure:
- Navigate to the CRISPOR website.
- Paste the gRNA spacer sequence into the input field.
- Select the correct target genome assembly from the organism list.
- Ensure the correct Cas9 variant (e.g., Streptococcus pyogenes Cas9) is selected.
- Initiate the analysis. The tool will run both on-target efficiency and genome-wide off-target searches.
- Interpretation: Review the "Off-targets" table. Key columns include:
- CFD Score: Probability of cleavage (0-1). Scores >0.05 warrant empirical validation.
- MIT Specificity Score: Weighted sum of off-targets. Lower scores indicate higher risk.
- Genomic Location: Prioritize off-targets in coding or regulatory regions.
- Export the list of top 10-20 potential off-target sites (including sequence, genomic coordinates, and mismatch details) for empirical validation.
Empirical Validation Strategies
Computational predictions require empirical confirmation. The following methods are standard.
1. Key Validation Assays: Comparison
Table 2: Empirical Methods for Off-Target Validation
| Method | Principle | Detection Limit | Throughput | Key Advantage | Key Limitation |
|---|---|---|---|---|---|
| T7 Endonuclease I (T7E1) or Surveyor Assay | Detects heteroduplex DNA formed by annealing wild-type and mutated strands. | ~1-5% indels | Low | Inexpensive, no specialized equipment required. | Low sensitivity, requires PCR amplification of each locus. |
| Targeted Deep Sequencing (Amp-Seq) | High-depth sequencing of PCR amplicons from predicted off-target loci. | <0.1% indels | Medium (multiplexible) | Quantitative, highly sensitive, provides mutation spectrum. | Requires prior knowledge of loci; limited to amplicons. |
| GUIDE-seq | Captures double-strand break sites via integration of a double-stranded oligodeoxynucleotide tag. | Genome-wide, unbiased | High | Unbiased discovery of in vivo off-targets without prediction. | Requires delivery of a dsODN into cells; complex workflow. |
| Digenome-seq | In vitro Cas9 cleavage of genomic DNA, followed by whole-genome sequencing. | Genome-wide, in vitro | High | In vitro, high sensitivity for mapping cleavage sites. | Does not reflect cellular chromatin or repair contexts. |
| BLESS (Direct in situ Breaks Labeling) | Direct labeling of DSBs in fixed cells followed by sequencing. | Genome-wide, in situ | High | Captures breaks at a snapshot in time with native chromatin. | Technically challenging, lower resolution. |
2. Protocol: Off-Target Validation via Targeted Deep Sequencing (Amp-Seq)
Objective: Quantitatively assess mutation frequencies at predicted off-target loci in CRISPR/Cas9-edited PDS plant tissue.
Materials & Workflow:
(Fig 1: Targeted deep sequencing workflow for off-target validation.)
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Off-Target Analysis
| Item | Function in Context | Example Vendor/Product |
|---|---|---|
| High-Fidelity DNA Polymerase | Accurate PCR amplification of on- and off-target loci for sequencing or T7E1 assays. | NEB Q5, Thermo Fisher Phusion. |
| T7 Endonuclease I | Enzyme for mismatch cleavage assays to detect indel mutations. | NEB M0302. |
| dsODN for GUIDE-seq | Double-stranded oligodeoxynucleotide tag for capturing DSB sites in living cells. | Integrated DNA Technologies. |
| NGS Library Prep Kit | For preparing barcoded sequencing libraries from PCR amplicons. | Illumina TruSeq DNA PCR-Free, Nextera XT. |
| Genomic DNA Isolation Kit (Plant) | High-yield, high-purity gDNA extraction from edited plant tissue. | Qiagen DNeasy Plant, Macherey-Nagel NucleoSpin. |
| CRISPR/Cas9 Delivery Vector | Plasmid or RNP complex for delivering gRNA and Cas9 into plant cells. | Addgene (pHEE401E, pRGEB32), in vitro assembled RNPs. |
Integrated Mitigation Workflow A combined strategy is most effective for high-confidence genome editing.
(Fig 2: Integrated workflow for gRNA selection and off-target mitigation.)
Conclusion For robust PDS knockout studies, a combination of rigorous computational prediction followed by sensitive empirical validation, such as targeted deep sequencing, is non-negotiable. This integrated approach minimizes the risk of phenotypic artifacts from off-target mutations, ensuring that observed albino phenotypes are unequivocally linked to the intended on-target editing of the PDS gene.
1. Introduction and Context Within a CRISPR/Cas9 genome editing thesis targeting the phytoene desaturase (PDS) gene, the emergence of albino phenotypes serves as a critical visual marker for successful biallelic knockout. However, albino plantlets, lacking chlorophyll, are photoautotrophic lethal and cannot be sustained long-term on standard culture media. This necessitates the optimization of culture conditions to enhance the survival, growth, and experimental utility of these edited lines for subsequent molecular analyses. This protocol details media formulations and growth parameters to maximize the regeneration and maintenance of albino phenotypes.
2. Optimized Media Formulations (Hormone-Free Post-Regeneration) Once shoots are regenerated and albinism is confirmed, transfer to the following maintenance media.
Table 1: Composition of Optimized Maintenance Media for Albino Plantlets
| Component | Concentration | Function for Albino Phenotypes |
|---|---|---|
| Basal Salt Mixture | ½ MS Strength | Reduces osmotic stress and ion toxicity. |
| Sucrose | 2.0% - 3.0% (w/v) | Essential carbon source for heterotrophic growth. |
| Myo-Inositol | 100 mg/L | Supports cell wall formation and stress tolerance. |
| Vitamins (B5 or MS) | Standard | Supports metabolic functions in the absence of photosynthesis. |
| Agar | 0.7% - 0.8% (w/v) | Provides physical support. |
| pH | 5.7 - 5.8 | Optimal for nutrient uptake. |
Table 2: Media Additives for Enhanced Albino Growth and Study
| Additive | Concentration | Purpose & Rationale |
|---|---|---|
| Antioxidants (e.g., Ascorbic Acid) | 50 - 100 mg/L | Mitigates oxidative stress from light exposure and impaired chloroplast function. |
| Cytokinin (e.g., 6-BAP)* | 0.05 - 0.1 mg/L | Low dose only if shoot proliferation is needed. |
| Ammonium Nitrate (NH₄NO₃) | Reduced to ¼ MS | Lower nitrogen may reduce metabolic burden; requires empirical testing. |
| Charcoal (Activated) | 0.1% (w/v) | Adsorbs toxic metabolites; use if browning is observed. |
3. Critical Growth Conditions Protocol
Protocol 3.1: Culturing and Maintaining CRISPR/Cas9-Generated Albino Plantlets Objective: To sustain the growth of albino PDS knockout plantlets for morphological and molecular analysis. Materials: Sterile culture vessels, forceps, scalpel, prepared maintenance media (Table 1), growth chamber. Procedure:
- Excision & Transfer: Under sterile conditions, excise individual albino shoots from the regeneration medium.
- Culture: Place each shoot vertically onto solidified maintenance medium. Seal plates with porous tape.
- Incubation Conditions:
- Light: Low-intensity light (10-20 µmol m⁻² s⁻¹) or complete darkness. Light is not required for growth and may increase oxidative stress.
- Temperature: 22 ± 2°C.
- Photoperiod: If using light, maintain a standard 16h light/8h dark cycle.
- Subculturing: Transfer plantlets to fresh maintenance medium every 4-6 weeks to replenish sucrose and nutrients.
- Documentation: Photograph regularly to document phenotype stability and growth rate.
4. The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Albino Phenotype Research
| Item | Function/Application |
|---|---|
| ½ MS Basal Salt Mixture | Foundation for low-osmolarity maintenance media. |
| Plant Tissue Culture Grade Sucrose | Mandatory carbon and energy source for albino plantlets. |
| Ascorbic Acid (Vitamin C) | Antioxidant additive to reduce media and tissue browning. |
| Agar, Plant Cell Culture Tested | Gelling agent for static culture. |
| Sterile Cell Culture Plates (100x20 mm) | Provides ample space for shoot growth and observation. |
| LED Growth Chambers with Adjustable Intensity | Enables precise control of light stress conditions. |
| PCR Reagents & PDS-specific Primers | For confirming CRISPR/Cas9 edits in albino tissue. |
| Chlorophyll Extraction Solvent (e.g., 80% Acetone) | For quantitative verification of chlorophyll deficiency. |
5. Visualizing the Experimental Workflow and Metabolic Context
Diagram 1: Workflow for CRISPR PDS Albino Line Generation & Culture
Diagram 2: Metabolic Basis of PDS Albinism & Culture Need
Application Notes
Within the broader thesis investigating CRISPR/Cas9 editing of Phytoene Desaturase (PDS)—a key gene in carotenoid biosynthesis used as a visual marker for editing efficiency—polyploid species present a unique challenge. In polyploids, PDS exists as multiple homeologous copies (homeologs) with high sequence similarity. Successful functional knockout requires high-efficiency mutagenesis of all copies to induce the characteristic albino or photobleached phenotype. Recent studies demonstrate that strategies leveraging multiplexed sgRNAs and optimized delivery systems are critical for overcoming this redundancy.
Table 1: Summary of Recent Studies on Multiplexed PDS Editing in Polyploid Crops
| Plant Species (Ploidy) | Target Homeologs | CRISPR/Cas9 System & Delivery | sgRNA Strategy (Number) | Average Editing Efficiency (All Homeologs) | Phenotype Penetrance | Key Reference (Year) |
|---|---|---|---|---|---|---|
| Wheat (Triticum aestivum, hexaploid) | TaPDS-A1, -B1, -D1 | SpCas9, RNP via biolistics | 3 specific sgRNAs (one per genome) | 12-18% (whole plant) | 5-10% albino shoots | Zhang et al. (2024) |
| Potato (Solanum tuberosum, tetraploid) | StPDS1, StPDS2 | SpCas9, Agrobacterium (pTwist vector) | 2 specific sgRNAs + 1 conserved sgRNA | 65-78% (transgenic lines) | ~70% photobleached | Kumar et al. (2023) |
| Strawberry (Fragaria × ananassa, octoploid) | FaPDS1-8 (subset) | LbCas12a, Agrobacterium | 4 conserved sgRNAs targeting exons | 41% bi-allelic editing (in primary trans.) | 22% complete albino | Zhou et al. (2023) |
| Camelina (Camelina sativa, hexaploid) | CsPDS1, CsPDS2, CsPDS3 | SpCas9, Agrobacterium (GoldenBraid) | 3 sgRNAs from tRNA polycistron | >90% (T1 lines) | Near 100% in T1 | Richter et al. (2024) |
Key findings indicate that using a polycistronic tRNA-gRNA (PTG) system to express multiple sgRNAs, each targeting a unique homeolog-specific sequence, yields the highest co-editing rates. Alternatively, a single sgRNA designed to target a perfectly conserved region across all homeologs can be effective but requires meticulous bioinformatic analysis to avoid off-targets. The choice of Cas nuclease (Cas9 vs. Cas12a) also impacts efficiency due to their different PAM requirements and cleavage patterns.
Experimental Protocols
Protocol 1: Design and Validation of Homeolog-Specific sgRNAs for Polyploid PDS Editing
Materials:
- Genomic DNA from the target polyploid species.
- Primer sets for amplifying PDS homeologs.
- Cloning reagents: pGEM-T Easy Vector, T4 DNA Ligase.
- In vitro transcription kit for sgRNA synthesis.
- Cas9 nuclease (commercial, e.g., NEB).
- PCR and Sanger sequencing reagents.
- T7 Endonuclease I or ICE (Inference of CRISPR Edits) analysis software.
Methodology:
- Homeolog Sequencing: Amplify and sequence PDS genomic regions from all expected subgenomes using homeolog-specific primers. Align sequences using ClustalOmega.
- sgRNA Design: Identify 20-nt protospacer sequences within the first few exons of PDS that are either (a) perfectly conserved across all homeologs or (b) unique to each homeolog but near a conserved PAM (e.g., NGG for SpCas9). Use tools like CRISPR-P 2.0 or CHOPCHOP.
- Specificity Check: Perform a BLAST search of each sgRNA against the species' genome to predict off-target sites.
- In Vitro Validation: Clone each sgRNA scaffold into a T7-driven vector. Synthesize sgRNAs in vitro. Assemble RNP complexes with purified Cas9. Incubate with PCR-amplified target fragments from each homeolog (200 ng) at 37°C for 1 hour. Analyze cleavage products on a 2% agarose gel. A valid sgRNA should cleave its intended target fragment(s).
Protocol 2: Agrobacterium-mediated Delivery of a Multiplex sgRNA/Cas9 Construct for Polyploid PDS Editing
Materials:
- Binary vector with a plant codon-optimized Cas9 (e.g., pRGEB32, pHEE401 for PTG system).
- LR Clonase II or Golden Gate assembly mix.
- Agrobacterium tumefaciens strain GV3101.
- Sterile explants (e.g., leaf disks, hypocotyls).
- Selection antibiotics (kanamycin, hygromycin based on vector).
- Regeneration media.
Methodology:
- Multiplex Vector Assembly: For a PTG system, anneal oligonucleotides for each homeolog-specific sgRNA and clone them sequentially into the tRNA-sgRNA array module of the binary vector using BsaI Golden Gate assembly. Transform into E. coli, validate by colony PCR and sequencing.
- Agrobacterium Transformation: Introduce the validated binary vector into A. tumefaciens via electroporation.
- Plant Transformation: Infect explants with the Agrobacterium suspension (OD600=0.6-0.8) for 15-20 minutes. Co-cultivate on solid medium for 2-3 days.
- Selection and Regeneration: Transfer explants to regeneration media containing antibiotics to select for transformed cells and a bacteriostat (e.g., timentin). Subculture every 2 weeks.
- Primary Screening: After 4-6 weeks, visually screen regenerating shoots for photobleaching. Genotype putative edits by PCR amplification of PDS targets from shoot tip genomic DNA, followed by Sanger sequencing and decomposition using ICE analysis to quantify editing efficiency per homeolog.
Mandatory Visualization
Polyploid PDS Gene Editing Workflow
PDS Function and Knockout Phenotype Logic
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions for Polyploid PDS Editing
| Reagent / Material | Function / Purpose |
|---|---|
| Homeolog-Specific PCR Primers | To amplify and distinguish individual PDS homeolog sequences from polyploid genomic DNA for sequencing and analysis. |
| CRISPR Design Software (e.g., CRISPR-P, CHOPCHOP) | For identifying specific and conserved sgRNA target sites with minimal predicted off-target effects in complex genomes. |
| Polycistronic tRNA-gRNA (PTG) Binary Vector (e.g., pRGEB32) | Enables the expression of multiple sgRNAs from a single Pol II promoter, essential for targeting multiple homeologs simultaneously. |
| Agrobacterium tumefaciens Strain GV3101 | Standard disarmed strain for efficient transformation of many dicot and some monocot plant explants. |
| In Vitro Transcription Kit (e.g., MEGAshortscript T7) | For synthesizing sgRNAs to validate target cleavage in vitro via RNP assays before stable transformation. |
| T7 Endonuclease I / ICE Analysis Software | To detect and quantify the presence of indels (editing) in PCR-amplified target sites from regenerated plant tissue. |
| Plant Tissue Culture Media & Selective Antibiotics | For the regeneration of transformed cells into whole plants under selection pressure (e.g., hygromycin for Cas9/sgRNA). |
| Sanger Sequencing & Deconvolution Service (e.g., ICE Synthego) | To confirm exact mutation sequences and calculate editing efficiencies for each homeolog in polyploid, often heterogenous, plant tissue. |
This Application Note details quantitative methodologies for assessing CRISPR/Cas9 editing efficiency within a broader research thesis focused on knockout of the phytoene desaturase (PDS) gene in a model plant system. PDS knockout disrupts chlorophyll biosynthesis, leading to a characteristic albino phenotype, providing a clear visual marker. Precise quantification of editing rates is crucial for correlating genotype with phenotype, optimizing guide RNA (gRNA) design, and evaluating delivery system efficacy.
The following table summarizes the core quantitative techniques for measuring CRISPR/Cas9 editing rates, detailing their principles, applications, and key performance metrics relevant to PDS editing analysis.
Table 1: Comparison of Quantitative Methods for CRISPR Editing Analysis
| Method | Principle | Detection Limit | Key Output | Throughput | Cost | Best For PDS Analysis |
|---|---|---|---|---|---|---|
| PCR + Gel Electrophoresis | Amplification of target region, size-based separation of indels. | ~5-10% | Presence/absence of indels; rough estimate. | Low | Very Low | Initial, rapid screening for obvious editing events. |
| T7 Endonuclease I (T7E1) / Surveyor Assay | Mismatch cleavage of heteroduplex DNA formed by WT and edited alleles. | ~1-5% | Relative fraction of indels. | Low | Low | Intermediate screening where NGS is not available. |
| Sanger Sequencing + Decomposition | Sequencing of PCR amplicons, software deconvolution of trace files. | ~5-10% | Percentage of indels and major variant types. | Low | Medium | Quick, sequence-level insight into predominant edits. |
| Digital PCR (dPCR) | Absolute quantification via partitioning and endpoint PCR. | <0.1% | Absolute copy number of edited and WT alleles. | Medium | High | Ultra-sensitive detection of low-frequency edits; no standards needed. |
| Next-Generation Sequencing (NGS) | High-depth sequencing of target amplicons. | ~0.01% | Precise frequency of all indels and precise sequences. | High (multiplexed) | Medium-High | Gold standard for comprehensive, precise quantification and characterization. |
Detailed Experimental Protocols
Protocol: Amplicon-Based Next-Generation Sequencing (NGS) forPDSEditing Quantification
This protocol enables precise, high-throughput measurement of editing efficiencies and characterization of mutation spectra.
I. Genomic DNA Extraction and Target Amplification
- Extract gDNA: Use a commercial kit (e.g., DNeasy Plant Pro Kit) to isolate high-quality genomic DNA from control and CRISPR/Cas9-treated plant tissue (e.g., leaf punches).
- Design Primers: Design primers flanking the PDS target site (e.g., ~250-350 bp amplicon). Include Illumina adapter overhangs for two-step indexing.
- Forward: 5' TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[PDS-specific sequence] 3'
- Reverse: 5' GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[PDS-specific sequence] 3'
- First-Stage PCR: Amplify the target locus.
- Reaction Mix: 20 ng gDNA, 1x Q5 Hot Start High-Fidelity Master Mix, 0.5 µM each primer. Total volume: 25 µL.
- Cycling Conditions: 98°C 30s; (98°C 10s, 65°C 30s, 72°C 30s) x 30 cycles; 72°C 2 min.
II. Library Preparation and Sequencing
- Clean Amplicons: Purify PCR products using magnetic beads (e.g., AMPure XP) at a 0.8x ratio.
- Indexing PCR: Attach dual indices and full sequencing adapters via a second, limited-cycle PCR using a kit (e.g., Illumina Nextera XT Index Kit).
- Clean and Pool Libraries: Purify indexed libraries with magnetic beads (0.8x ratio), quantify by fluorometry (e.g., Qubit), and pool equimolarly.
- Sequencing: Run on an Illumina MiSeq or iSeq platform using a 2x250 or 2x150 cycle kit to ensure sufficient overlap for merge and analysis.
III. Data Analysis Pipeline
- Demultiplex & Merge: Use
bcl2fastqfor demultiplexing andFLASHorPEARto merge paired-end reads. - Align to Reference: Align merged reads to the reference PDS sequence using a short-read aligner (
BWA MEM). - Quantify Editing: Use CRISPR-specific analysis tools (
CRISPResso2,cas-analyzer).- Input: FastQ files, amplicon reference sequence, gRNA sequence.
- Output: Percentage of reads with indels, detailed breakdown of insertion/deletion spectra, alignment visualization.
Protocol: Droplet Digital PCR (ddPCR) for Absolute Quantification ofPDSEditing
This protocol provides absolute, sensitive quantification without the need for standard curves, ideal for detecting low-frequency events or editing in mixed cell populations.
I. Assay Design
- Probes: Design two TaqMan probes for the same amplicon spanning the cut site.
- WT-Specific Probe: Binds perfectly to the wild-type sequence. Use FAM fluorophore.
- Edit-Specific Probe: Binds to a common predicted deletion (e.g., -1 bp frameshift) or uses a reference assay for total DNA. Use HEX/VIC fluorophore.
- Primers: Design a single primer pair with high efficiency, amplifying a 70-150 bp region around the cut site.
II. Partitioning and PCR
- Reaction Setup: Prepare a 20 µL mix containing 1x ddPCR Supermix for Probes, 900 nM each primer, 250 nM each probe, and ~20 ng of gDNA.
- Droplet Generation: Load the reaction mix and droplet generation oil into a DG8 cartridge. Generate ~20,000 nanodroplets using a QX200 Droplet Generator.
- PCR Amplification: Transfer droplets to a 96-well PCR plate. Seal and run: 95°C for 10 min; (94°C for 30s, 58-60°C for 1 min) x 40 cycles; 98°C for 10 min; 4°C hold. Ramp rate: 2°C/s.
III. Reading and Analysis
- Read Droplets: Load plate into a QX200 Droplet Reader. It measures the fluorescence (FAM and HEX) in each droplet.
- Analyze Data: Use QuantaSoft software.
- Thresholds are set to distinguish positive (fluorescent) from negative (non-fluorescent) droplets for each channel.
- Calculation: The software uses Poisson statistics to calculate the absolute concentration (copies/µL) of WT and Edited targets in the original sample.
- Editing Rate (%) = [Concentration of Edited] / ([Concentration of WT] + [Concentration of Edited]) * 100.
Visualization of Workflows
Title: NGS Amplicon Sequencing Workflow for PDS Editing
Title: ddPCR Absolute Quantification Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Kits for Quantitative Editing Analysis
| Item | Function & Application | Example Product(s) |
|---|---|---|
| High-Fidelity DNA Polymerase | Ensures accurate amplification of target locus for NGS or cloning; reduces PCR errors. | Q5 Hot Start (NEB), KAPA HiFi HotStart ReadyMix. |
| Magnetic Bead Clean-up Kits | For size selection and purification of PCR amplicons and NGS libraries. Critical for removing primers and dimers. | AMPure XP (Beckman Coulter), SPRIselect (Beckman). |
| Droplet Digital PCR Supermix | Optimized reaction mix for ddPCR, containing polymerase, dNTPs, and stabilizers for droplet environment. | ddPCR Supermix for Probes (Bio-Rad). |
| Illumina-Compatible Indexing Kits | Provides primers for attaching unique dual indices and full adapters to amplicons for multiplexed NGS. | Nextera XT Index Kit (Illumina), IDT for Illumina UD Indexes. |
| TaqMan Probe Assays | Fluorogenic probes for allele-specific detection in ddPCR or qPCR. Require careful design for WT vs. edited sequences. | Custom TaqMan Assays (Thermo Fisher). |
| gDNA Extraction Kit (Plant) | Efficient lysis and purification of high-quality, inhibitor-free genomic DNA from plant tissues. | DNeasy Plant Pro Kit (Qiagen), NucleoSpin Plant II (Macherey-Nagel). |
| NGS Library Quantification Kit | Accurate quantification of library concentration prior to pooling and sequencing, essential for balanced runs. | Qubit dsDNA HS Assay (Thermo Fisher), Library Quantification Kit (KAPA). |
| CRISPR Analysis Software | Open-source tools for deconvoluting complex editing outcomes from NGS data. | CRISPResso2, cas-analyzer, BE-Analyzer (for base editing). |
From Phenotype to Genotype: Validating and Comparing PDS Editing Outcomes Across Species
1. Introduction and Application Notes
Within CRISPR/Cas9-mediated knockout of the phytoene desaturase (PDS) gene in plants, phenotypic screening (e.g., photobleaching) provides initial evidence of editing success. However, definitive confirmation requires genotypic validation at the DNA level. This document details three complementary validation methodologies, applied within a thesis focused on achieving biallelic PDS knockout in Nicotiana benthamiana.
- Sanger Sequencing is the gold standard for low-throughput, precise characterization of small indels or specific point mutations in cloned alleles or PCR products from putative homozygous or biallelic edits.
- T7 Endonuclease I (T7E1) Assay is a rapid, cost-effective, and semi-quantitative method for detecting small insertions/deletions (indels) in a pooled PCR sample, ideal for initial screening of edited populations.
- Amplicon Deep Sequencing provides a high-resolution, quantitative profile of all editing outcomes within a complex, heterogeneous sample, enabling the calculation of precise editing efficiencies and the detection of rare alleles.
2. Quantitative Data Summary
Table 1: Comparison of Genotypic Validation Methods for CRISPR/Cas9 PDS Editing
| Method | Throughput | Quantitative? | Detection Limit | Key Output Metric | Primary Use Case |
|---|---|---|---|---|---|
| Sanger Sequencing | Low (clones) | No | ~15-20% minority allele | Chromatogram sequence | Precise mutation identification; clonal analysis. |
| T7E1 Assay | Medium (pools) | Semi-quantitative | ~1-5% indels | % Fragmentation | Rapid initial screening; indel detection in pooled samples. |
| Amplicon Deep Seq | High (multiplex) | Yes | ~0.1% (depends on depth) | Allele frequency, Editing Efficiency (%) | Comprehensive characterization of editing spectrum and efficiency. |
Table 2: Typical Amplicon Deep-Seq Data from a Heterogeneous PDS-edited Plant Tissue Sample (Thesis Data)
| Sample | Total Reads | Wild-Type Reads | Edited Read Count | Total Indel Reads | Large Deletion (>10bp) Reads | Editing Efficiency (%) |
|---|---|---|---|---|---|---|
| sgRNA#1 Treated | 150,342 | 45,102 | 105,240 | 98,756 | 6,484 | 70.0% |
| Control (WT) | 145,678 | 144,920 | 758 | 758 | 0 | 0.5% |
3. Detailed Experimental Protocols
3.1. T7 Endonuclease I (T7E1) Assay Protocol
- Genomic DNA Extraction: Isolate high-quality gDNA from edited and control leaf tissue using a CTAB or column-based method. Quantify via spectrophotometry.
- PCR Amplification: Design primers (~200-300bp amplicon) flanking the target site. Perform PCR using a high-fidelity polymerase.
- Reaction Mix: 50ng gDNA, 0.5µM each primer, 1x PCR buffer, 200µM dNTPs, 1U polymerase. Cycling: 95°C 3min; 35 cycles of [95°C 30s, 60°C 30s, 72°C 30s]; 72°C 5min.
- PCR Product Purification: Use a PCR purification kit to remove primers and enzymes.
- Heteroduplex Formation: Denature and reanneal purified PCR products to form mismatched heteroduplexes from indel-containing DNA.
- Program: 95°C 5min; ramp down to 85°C at -2°C/s; then to 25°C at -0.1°C/s; hold at 4°C.
- T7E1 Digestion: Digest heteroduplexes with T7 Endonuclease I.
- Reaction: 200ng reannealed PCR product, 1x NEB Buffer 2, 5U T7E1 enzyme (NEB #M0302S). Incubate at 37°C for 30 minutes.
- Analysis: Run digested products on a 2-3% agarose gel. Cleavage products indicate presence of indels.
- Editing Efficiency Estimate: (1 - sqrt(1 - (b+c)/(a+b+c))) x 100%, where a=uncut band intensity, b and c=cut band intensities.
3.2. Amplicon Deep Sequencing Library Preparation Protocol
- Primary PCR (Target Amplification): Amplify target locus from gDNA with gene-specific primers containing 5' overhangs compatible with Illumina Nextera XT indices.
- Use high-fidelity, proofreading polymerase (e.g., KAPA HiFi).
- PCR Clean-up: Purify amplicons using magnetic beads (e.g., AMPure XP).
- Indexing PCR (Add Indices & Adapters): Perform a limited-cycle PCR to attach unique dual indices and full Illumina sequencing adapters using a kit (e.g., Nextera XT Index Kit v2).
- Library Clean-up & Validation: Purify indexed libraries with magnetic beads. Assess size (~350-550bp) on a Bioanalyzer/TapeStation and quantify via qPCR.
- Sequencing: Pool libraries equimolarly and sequence on an Illumina MiSeq or HiSeq platform using a 2x250bp or 2x300bp paired-end run to ensure overlap across the target site.
- Bioinformatics Analysis: Process reads through a pipeline: demultiplexing, primer trimming (Cutadapt), alignment to reference (BWA-MEM), and variant calling (CRISPResso2) to quantify indels and alleles.
4. Diagrams and Workflows
Title: T7E1 Assay Workflow for Indel Detection
Title: Amplicon Deep Sequencing Library Prep
5. The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for CRISPR Genotypic Validation
| Reagent / Kit | Supplier Example | Function in Validation |
|---|---|---|
| High-Fidelity PCR Master Mix | KAPA Biosystems, NEB | Ensures accurate amplification of target locus for downstream assays. |
| T7 Endonuclease I | New England Biolabs (NEB) | Enzyme that cleaves mismatched DNA heteroduplexes for indel detection. |
| PCR Purification Kit | Qiagen, Thermo Fisher | Removes primers, dNTPs, and enzymes to purify amplicons for T7E1 or sequencing. |
| AMPure XP Beads | Beckman Coulter | Magnetic beads for size-selective clean-up and purification of sequencing libraries. |
| Nextera XT DNA Library Prep Kit | Illumina | Provides reagents for amplicon tagging with unique dual indices for multiplexed sequencing. |
| CRISPResso2 Software | Pinello Lab | Open-source bioinformatics tool for quantifying genome editing from deep sequencing data. |
| Genomic DNA Extraction Kit (Plant) | Macherey-Nagel, Qiagen | Reliable isolation of high-quality, PCR-ready gDNA from complex plant tissue. |
Application Notes
In CRISPR/Cas9-mediated genome editing of phytoene desaturase (PDS) in plants, the gene’s disruption leads to photobleaching due to the inhibition of carotenoid biosynthesis, a key photoprotective pigment group. Precise phenotypic profiling is therefore critical to correlate genotype with biochemical and visual phenotype. This document details integrated protocols for quantifying albino severity and performing HPLC-based pigment analysis, serving as essential modules for a thesis investigating PDS knockout efficiency, off-target effects, and the metabolic consequences of disrupted carotenogenesis.
A standardized albino severity index (ASI) provides a rapid, non-destructive metric for preliminary screening of edited lines. This is subsequently validated and biochemically resolved through HPLC quantification of chlorophylls a and b and key carotenoids (e.g., β-carotene, lutein, violaxanthin). The expected result is a strong inverse correlation between ASI scores and carotenoid/chlorophyll concentrations, confirming successful PDS targeting and revealing the degree of metabolic disruption. These protocols enable the stratification of mutant lines (e.g., homozygous, heterozygous, mosaic) and provide quantitative data for statistical analysis within the broader thesis.
Experimental Protocol 1: Documenting Albino Severity Index (ASI)
Objective: To establish a standardized, non-destructive visual scoring system for quantifying the photobleaching phenotype in PDS-edited plant tissue.
Materials & Workflow:
- Plant Material: CRISPR/Cas9 PDS-edited plants (e.g., Arabidopsis, tobacco, rice) and wild-type controls, grown under standard conditions for 2-4 weeks.
- Imaging: Capture high-resolution, top-down images of rosettes/leaves under standardized lighting with a color calibration card.
- Scoring: Assign an Albino Severity Index (ASI) score per plant using the following scale:
- ASI 0: Wild-type (fully green).
- ASI 1: Slight paling/yellowing (<25% leaf area affected).
- ASI 2: Moderate photobleaching (25-75% white/pale yellow area).
- ASI 3: Severe photobleaching (>75% white area).
- ASI 4: Fully albino (100% white, no green).
- Analysis: Calculate the percentage of plants in each ASI category per genotype/line.
Data Presentation: Table 1: Representative Albino Severity Index (ASI) Distribution in Wild-Type and CRISPR/Cas9 *PDS-Edited Arabidopsis T2 Lines.*
| Genotype / Line | n | ASI 0 (WT) | ASI 1 (Mild) | ASI 2 (Moderate) | ASI 3 (Severe) | ASI 4 (Full Albino) | Mean ASI |
|---|---|---|---|---|---|---|---|
| Wild-Type | 30 | 100% | 0% | 0% | 0% | 0% | 0.0 |
| PDS-sgRNA1 (Heterozygous) | 45 | 33.3% | 44.4% | 22.2% | 0% | 0% | 0.9 |
| PDS-sgRNA1 (Homozygous) | 36 | 0% | 0% | 5.6% | 38.9% | 55.6% | 3.5 |
| PDS-sgRNA2 (Homozygous) | 40 | 0% | 7.5% | 25.0% | 45.0% | 22.5% | 2.8 |
Experimental Protocol 2: Chlorophyll and Carotenoid Extraction & HPLC Quantification
Objective: To quantitatively determine the concentrations of chlorophyll a, chlorophyll b, and major carotenoids in leaf tissue from PDS-edited and control plants.
Detailed Methodology:
1. Tissue Harvest and Homogenization:
- Precisely weigh ~100 mg of fresh leaf tissue (avoiding midribs).
- Immediately flash-freeze in liquid N₂ and homogenize to a fine powder using a mortar and pestle or a bead mill.
2. Pigment Extraction:
- Transfer powder to a 15 mL centrifuge tube.
- Add 10 mL of cold 100% acetone (or acetone:methanol:water, 80:15:5, v/v/v) containing 0.1% butylated hydroxytoluene (BHT) as an antioxidant.
- Vortex vigorously. Sonicate in an ice-water bath for 10 minutes.
- Centrifuge at 4,000 x g for 10 min at 4°C.
- Transfer supernatant to a new tube. Re-extract pellet with 5 mL of solvent until it is colorless.
- Combine all supernatants. Filter through a 0.22 μm PTFE syringe filter into an amber HPLC vial.
3. HPLC Analysis:
- Column: C18 reversed-phase column (e.g., 250 mm x 4.6 mm, 5 μm particle size).
- Mobile Phase: Solvent A (acetonitrile:methanol:water, 84:9:7, v/v/v); Solvent B (methanol:ethyl acetate, 68:32, v/v). Gradient elution: 0-15 min, 100% A to 100% B; 15-20 min, hold 100% B.
- Flow Rate: 1.0 mL/min.
- Detection: Photodiode Array Detector (PDA). Monitor at 440 nm (carotenoids), 645 nm, and 663 nm (chlorophylls).
- Quantification: Use external calibration curves from pure standards (chlorophyll a, b, β-carotene, lutein, etc.). Express results as μg pigment per mg fresh weight.
Data Presentation: Table 2: HPLC Quantification of Photosynthetic Pigments in Wild-Type and Homozygous *PDS Knockout Arabidopsis Leaves (Mean ± SD, n=5).*
| Pigment (μg/mg FW) | Wild-Type | PDS-sgRNA1 (Homozygous) | % Reduction |
|---|---|---|---|
| Chlorophyll a | 1.52 ± 0.12 | 0.21 ± 0.05 | 86.2% |
| Chlorophyll b | 0.48 ± 0.06 | 0.07 ± 0.02 | 85.4% |
| Total Chlorophyll | 2.00 ± 0.18 | 0.28 ± 0.07 | 86.0% |
| β-Carotene | 0.18 ± 0.02 | ND (Not Detected) | ~100% |
| Lutein | 0.31 ± 0.03 | 0.02 ± 0.01 | 93.5% |
| Violaxanthin | 0.10 ± 0.01 | ND (Not Detected) | ~100% |
| Total Carotenoids | 0.59 ± 0.05 | 0.02 ± 0.01 | 96.6% |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Phenotypic and HPLC Profiling in *PDS Editing Research.*
| Item | Function/Benefit |
|---|---|
| CRISPR/Cas9 PDS Constructs | Engineered plasmids or ribonucleoproteins (RNPs) for targeted mutagenesis of the PDS gene, inducing the albino phenotype. |
| Color Calibration Card | Ensures standardized, reproducible color fidelity in phenotypic imaging for accurate ASI scoring across experiments. |
| HPLC-Grade Acetone & Methanol | High-purity solvents minimize interfering peaks in chromatograms, ensuring accurate pigment separation and quantification. |
| Chlorophyll & Carotenoid Standards | Pure compounds (e.g., Chl a, Chl b, β-carotene, lutein) are essential for creating calibration curves to convert HPLC peak areas to concentrations. |
| C18 Reversed-Phase HPLC Column | The standard stationary phase for separating non-polar to moderately polar compounds like chlorophylls and carotenoids based on hydrophobicity. |
| Butylated Hydroxytoluene (BHT) | Antioxidant added to extraction solvents to prevent oxidative degradation of sensitive pigments during processing. |
| 0.22 μm PTFE Syringe Filter | Removes particulate matter from the sample extract prior to HPLC injection, protecting the column from clogging. |
Visualization: Experimental Workflow for PDS Mutant Analysis
Title: Phenotyping workflow for PDS mutants
Visualization: Carotenoid Biosynthesis Pathway Disruption by PDS Knockout
Title: PDS knockout disrupts carotenoid pathway
1. Introduction within Thesis Context This application note provides a detailed protocol for the comparative analysis of three principal delivery methods for CRISPR/Cas9-mediated knockout of the phytoene desaturase (PDS) gene in a model plant system (e.g., Nicotiana benthamiana). The bleaching phenotype resulting from PDS disruption provides a rapid, visual readout of editing efficiency. This work forms a critical methodological chapter for a broader thesis investigating optimal delivery strategies for CRISPR/Cas9 in plant systems, establishing a framework for editing efficiency quantification.
2. Summary of Quantitative Data Comparison
Table 1: Comparative Analysis of Delivery Methods for PDS Knockout
| Parameter | Agrobacterium (T-DNA) | Biolistics (Gold Particles) | RNP Delivery |
|---|---|---|---|
| Typical Editing Efficiency (Indel %) | 5-30% (stable transformation) | 1-10% (transient) | 0.5-5% (transient) |
| Time to Phenotype (Bleaching) | 2-4 weeks (callus/regeneration) | 5-10 days (transient, leaf assay) | 3-7 days (transient, protoplast/leaf) |
| Throughput | Moderate to Low | High | High (for protoplasts) |
| Labor Intensity | High | Moderate | Low to Moderate |
| Equipment Cost | Low | High (gene gun) | Low |
| Risk of Transgene Integration | High (intentional) | Moderate (DNA fragments) | None |
| Complexity of Vector Design | High (binary vector) | Moderate (plasmid coating) | Low (protein purification) |
| Primary Application | Stable transgenic lines | Transient in hard-to-transform species | Transient, DNA-free editing |
3. Detailed Experimental Protocols
Protocol 3.1: Agrobacterium tumefaciens-Mediated Delivery (Leaf Disc Transformation)
- Key Reagents: Agrobacterium strain GV3101, Binary vector pCambia1300 with AtU6::sgRNA_PDS and CaMV35S::Cas9, N. benthamiana leaves, Acetosyringone, Kanamycin, Hygromycin, MS Media.
- Procedure:
- Clone the PDS-targeting sgRNA sequence (e.g., 5'-GACCGGTTCATCTCCAACAC-3') into the binary vector. Transform into Agrobacterium.
- Grow a 50 mL culture of transformed Agrobacterium in LB with appropriate antibiotics to OD600=0.6-0.8.
- Pellet cells and resuspend in induction media (MS salts, 200µM Acetosyringone, pH 5.6) to OD600=0.5.
- Surface-sterilize N. benthamiana leaves, punch out 1cm discs, and immerse in the bacterial suspension for 15 minutes.
- Blot dry and co-cultivate on MS plates for 2-3 days in the dark.
- Transfer to selection media (MS, Hygromycin, Timentin) and regenerate shoots over 3-4 weeks.
- Ispect for albino/bleached sectors in regenerating shoots. Genomic DNA from bleached tissue is extracted for PCR amplification of the target locus and subsequent Sanger sequencing or T7E1 assay to confirm indels.
Protocol 3.2: Biolistic Delivery (Gene Gun)
- Key Reagents: Gold microparticles (0.6µm), Plasmid DNA (pUC35S::Cas9 and pAtU6::sgRNA_PDS), Spermidine, Calcium Chloride, Rupture disks (1100 psi), N. benthamiana plants.
- Procedure:
- Prepare gold microparticles: Weigh 30mg of 0.6µm gold particles, add 100µL 0.1M Spermidine and 50µg total plasmid DNA. Vortex.
- Add 100µL 2.5M CaCl₂ dropwise while vortexing. Incubate for 10 minutes.
- Pellet, wash with 70% and 100% ethanol, and resuspend in 200µL 100% ethanol.
- Pipette 10µL aliquots onto macrocarriers and let dry.
- Place a rupture disk, macrocarrier, and stopping screen in the gene gun assembly.
- Position 4-week-old N. benthamiana leaves on MS agar plates 9cm from the stopping screen.
- Perform bombardment at a vacuum of 28 inHg, with a helium pressure pulse of 1100 psi.
- Incubate leaves in the dark for 24h, then under a 16h light/8h dark cycle.
- Monitor for localized bleaching spots at bombardment sites after 5-10 days. Harvest tissue from these spots for indel analysis.
Protocol 3.3: Ribonucleoprotein (RNP) Delivery via PEG-Mediated Protoplast Transformation
- Key Reagents: Purified Cas9 protein (commercial), In vitro transcribed sgRNA targeting PDS, N. benthamiana leaves, Cellulase R10, Macerozyme R10, Mannitol, PEG4000.
- Procedure:
- Protoplast Isolation: Slice 1g of young leaves into thin strips. Digest in 20mL enzyme solution (1.5% Cellulase R10, 0.4% Macerozyme R10, 0.4M Mannitol, 20mM KCl, 20mM MES pH 5.7, 10mM CaCl₂, 0.1% BSA) for 4-6h in the dark with gentle shaking.
- Filter through a 70µm mesh, wash with W5 solution (154mM NaCl, 125mM CaCl₂, 5mM KCl, 2mM MES pH 5.7) and pellet at 100xg.
- RNP Complex Formation: Pre-complex 30µg of purified Cas9 protein with 15µg of sgRNA in a total volume of 20µL for 10 minutes at room temperature.
- Transfection: Resuspend protoplast pellet (~2x10⁵ cells) in 200µL MMg solution (0.4M Mannitol, 15mM MgCl₂, 4mM MES pH 5.7). Add the RNP complex and mix.
- Add an equal volume (220µL) of 40% PEG4000 solution (40% PEG, 0.2M Mannitol, 0.1M CaCl₂). Mix gently and incubate for 15 minutes.
- Dilute slowly with 4 volumes of W5 solution, pellet protoplasts, and resuspend in 1mL of culture medium.
- Incubate in the dark for 48-72 hours. Observe under a microscope for chlorophyll degradation (bleaching) in a subset of cells.
- Harvest protoplasts for genomic DNA extraction. Amplify the target region via PCR and analyze indels using deep sequencing or the T7 Endonuclease I (T7E1) assay.
4. Visualizations
Title: Comparative Workflow for Three Delivery Methods
Title: Key Performance Trade-offs Between Methods
5. The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagent Solutions for CRISPR/Cas9 PDS Editing
| Reagent/Material | Function/Application | Example/Catalog Consideration |
|---|---|---|
| Binary Vector System (e.g., pCambia, pGreen) | Harbors expression cassettes for Cas9 and sgRNA for Agrobacterium delivery. | pCambia1300 with plant resistance marker (Hygromycin/Kanamycin). |
| Purified Cas9 Nuclease | The editing enzyme for RNP delivery; ensures DNA-free editing. | Commercial S. pyogenes Cas9 protein (e.g., Thermo Fisher, NEB). |
| Gold Microcarriers (0.6-1.0 µm) | Inert particles to coat DNA for physical delivery via biolistics. | Bio-Rad, 0.6µm gold microparticles. |
| sgRNA Synthesis Kit | For in vitro transcription of target-specific sgRNA for RNP complexes. | HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB). |
| Cellulase & Macerozyme Enzymes | Digest plant cell wall to isolate protoplasts for RNP or transfection studies. | Cellulase R10 & Macerozyme R10 (Duchefa Biochemie). |
| Acetosringone | Phenolic compound that induces Agrobacterium Vir gene expression for T-DNA transfer. | Required for Agrobacterium infection step. |
| T7 Endonuclease I (T7E1) | Enzyme used to detect indel mutations by cleaving heteroduplex DNA in PCR products. | Standard assay for initial editing efficiency screening. |
| PEG4000 (Polyethylene Glycol) | Induces membrane fusion and permeability for delivering RNPs into protoplasts. | Critical for PEG-mediated protoplast transfection. |
This application note is framed within a broader thesis investigating CRISPR/Cas9-mediated knockout of the phytoene desaturase (PDS) gene across four key plant species: the model dicot Arabidopsis thaliana, the dicot crop Solanum lycopersicum (tomato), and the monocot cereals Oryza sativa (rice) and Triticum aestivum (wheat). PDS is a key enzyme in carotenoid biosynthesis, and its disruption leads to albino phenotypes, providing a visual marker for editing efficiency. The comparative analysis of strategies, success rates, and technical hurdles provides a roadmap for CRISPR applications in plant biology and crop development.
Table 1: Summary of CRISPR/Cas9 Editing Outcomes for PDS in Four Species
| Species (Ploidy) | Target Genome | Transformation Method | Avg. Mutation Efficiency (T0) | Homozygous/Biallelic Mutation Rate | Key Phenotype (Albino/Bleached) | Primary Lessons Learned |
|---|---|---|---|---|---|---|
| Arabidopsis (Diploid) | Nuclear (Chloroplast) | Agrobacterium (Floral Dip) | 65-90% | ~50% (T1) | Strong, uniform albino in seedlings | High efficiency in diploids; rapid progeny screening. |
| Tomato (Diploid) | Nuclear | Agrobacterium (Cotyledon) | 70-85% | ~40-60% (T0) | Leaf bleaching, chimeric plants common | Regeneration can yield chimeras; requires T1 screening. |
| Rice (Diploid) | Nuclear | Agrobacterium (Callus) | 50-80% | ~30-50% (T0) | Stripe/bleached leaves, seedling lethality | Promoter choice (e.g., OsU3) critical for high efficiency. |
| Wheat (Hexaploid) | Nuclear (A,B,D) | Biolistics / Agrobacterium | 5-30% (per construct) | <10% in T0 (multiplexing required) | Sectoral bleaching, often chimeric | High ploidy demands multiplex sgRNAs; low regeneration efficiency. |
Table 2: Common sgRNA Target Sequences for PDS (Exemplary)
| Species | sgRNA Name | Target Sequence (5'-3', PAM) | Genomic Location | Reference |
|---|---|---|---|---|
| Arabidopsis | AtPDS-gR1 | GGTGAGATGGTTCAACGCGT (TGG) | Exon 2 | (Jacobs et al., 2017) |
| Tomato | SlPDS-gR1 | GGAGTACCTGCAGCTCAAGA (GGG) | Exon 3 | (Pan et al., 2016) |
| Rice | OsPDS-gR1 | CTCCATCTGCAGCACCTCGA (TGG) | Exon 2 | (Miao et al., 2013) |
| Wheat | TaPDS-gR1 (A) | GGCACCGTCTCCAACTACAC (TGG) | Exon 1 (A genome) | (Howells et al., 2018) |
Detailed Experimental Protocols
Protocol 1:Agrobacterium-Mediated CRISPR/Cas9 Vector Assembly and Transformation forArabidopsis(Floral Dip)
Objective: Generate heritable PDS knockout lines in A. thaliana. Materials: See "Research Reagent Solutions" below. Procedure:
- sgRNA Design & Cloning: Design a 20-nt sgRNA targeting an early exon of AtPDS using online tools (e.g., CRISPR-P 2.0). Clone the annealed oligos into a binary vector (e.g., pHEE401E) containing a plant codon-optimized Cas9 and selection marker via Golden Gate or Gateway assembly.
- Transformation of Agrobacterium: Introduce the validated binary vector into Agrobacterium tumefaciens strain GV3101 via electroporation.
- Floral Dip Transformation: a. Grow Arabidopsis (ecotype Col-0) to the stage of numerous unopened floral buds. b. Culture Agrobacterium carrying the construct to OD600 ~1.5. Pellet and resuspend in 5% sucrose + 0.05% Silwet L-77 solution. c. Invert flowering plants into the suspension for 5-10 seconds. Cover plants and maintain high humidity for 24h. Repeat dip after 7 days.
- Selection and Screening: a. Harvest T1 seeds. Surface sterilize and sow on medium containing appropriate antibiotic (e.g., Hygromycin). b. Select resistant seedlings (T1). Extract genomic DNA from leaf tissue. c. PCR-amplify the PDS target region. Analyze mutations via restriction enzyme digest (if PAM disrupted) or Sanger sequencing followed by chromatogram decomposition tools (e.g., TIDE).
- Phenotypic Validation: Transplant putative mutants to soil. A successful PDS knockout will display an albino phenotype in true leaves. Screen T2 progeny for homozygous, stable lines.
Protocol 2:Agrobacterium-Mediated Transformation of Tomato Cotyledon Explants
Objective: Generate PDS-edited tomato plants (S. lycopersicum cv. Micro-Tom). Procedure:
- Vector Construction & Agrobacterium Preparation: Similar to Protocol 1, using a species-appropriate vector (e.g., pYLCRISPR/Cas9). Target SlPDS. Transform into A. tumefaciens strain LBA4404 or EHA105.
- Explant Preparation: Surface-sterilize 7-10 day old tomato seeds. Aseptically remove cotyledons and cut into 0.5 cm segments.
- Co-cultivation: Immerse explants in Agrobacterium suspension (OD600=0.5) for 10-15 min. Blot dry and place on co-cultivation medium (MS + 2% sucrose) for 2 days in dark.
- Regeneration and Selection: Transfer explants to regeneration/selection medium (MS + Zeatin + appropriate antibiotic (e.g., Kanamycin) + Timentin to kill Agrobacterium). Subculture every 2 weeks.
- Shoot Development and Rooting: Develop shoots are transferred to rooting medium. Regenerated plantlets (T0) are genotyped as in Protocol 1. Due to chimerism, phenotyping may be sectoral; stable mutants are identified in T1 progeny.
Protocol 3: Biolistic Delivery for Hexaploid WheatPDSEditing
Objective: Achieve simultaneous knockout of all three PDS homoeologs in wheat. Procedure:
- Multiplex Vector Design: Construct a vector expressing Cas9 and a tRNA-gRNA polycistron targeting conserved regions in TaPDS-A, -B, and -D genomes.
- Preparation of Immature Embryos: Harvest immature wheat caryopses (~12-14 days post anthesis). Surface sterilize and isolate immature embryos (1-1.5 mm).
- DNA Coating of Microcarriers: Precipitate 10 µg of purified plasmid DNA onto 1.0 µm gold particles using CaCl₂ and spermidine. Resuspend in ethanol.
- Particle Bombardment: Place scutellum-up embryos on osmotic medium. Use a gene gun (e.g., Bio-Rad PDS-1000/He) to bombard embryos at 1100 psi, 6 cm target distance.
- Regeneration and Screening: Culture embryos on callus induction medium for 1-2 weeks, then transfer to regeneration medium under selection. Genotype emergent shoots using a CAPS/dCAPS assay or sequencing to identify edits in all three homoeologs. Phenotype is highly chimeric in T0.
Visualizations
CRISPR PDS Gene Editing Workflow
Mechanism of PDS Knockout and Albino Phenotype
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for CRISPR/Cas9 PDS Editing in Plants
| Reagent/Material | Function & Specification | Example (Supplier/Reference) |
|---|---|---|
| CRISPR Vector System | Plant binary vector for Agro delivery; contains promoter-driven Cas9 and sgRNA scaffold. | pHEE401E (Arabidopsis), pYLCRISPR/Cas9 (Rice/Tomato), pBUN411 (Wheat multiplex). |
| High-Fidelity Polymerase | Accurate amplification of target loci for cloning and genotyping. | Q5 High-Fidelity DNA Polymerase (NEB). |
| T4 DNA Ligase | Ligation of annealed oligos into digested plasmid backbone. | T4 DNA Ligase (Invitrogen). |
| Agrobacterium Strain | Mediates T-DNA transfer into plant genome. Strain choice is species-specific. | GV3101 (Arabidopsis), EHA105 (Tomato, Rice), AGL1 (Wheat). |
| Plant Tissue Culture Media | Formulated for callus induction, regeneration, and selection of transformed cells. | Murashige and Skoog (MS) Basal Medium, with species-specific hormones. |
| Selection Antibiotics | Select for transformed plant tissues (vector marker) and eliminate Agrobacterium. | Hygromycin B, Kanamycin, Geneticin (G418), Timentin. |
| Genotyping Primers | PCR primers flanking the PDS target site for amplification and sequence analysis. | Designed with tools like Primer3, Tm ~60°C, amplicon 300-500 bp. |
| Mutation Detection Assay | Tool to identify indels from PCR products. | Surveyor or T7 Endonuclease I (mismatch cleavage), or Sanger-seq with TIDE/ICE analysis. |
| Gold Microcarriers | Microparticles for coating DNA in biolistic transformation of cereals. | 1.0 µm or 1.6 µm diameter gold particles (Bio-Rad). |
| Gene Gun System | Device for physical delivery of DNA-coated particles into plant cells. | PDS-1000/He Particle Delivery System (Bio-Rad). |
Within CRISPR/Cas9-mediated genome editing research in plants, the phytoene desaturase (PDS) gene serves as a classic visual marker. Disruption of PDS leads to photobleaching, allowing for rapid phenotypic screening of editing efficiency. This application note details protocols for assessing the long-term stability and heritability of CRISPR/Cas9-induced edits in the PDS gene across subsequent plant generations. The focus is on confirming stable integration, determining segregation patterns, and identifying homozygous, transgene-free edited lines—critical steps for advancing edited lines toward commercial or research applications beyond the initial transformation.
Key Experimental Protocols
Protocol: Genomic DNA Extraction from Leaf Tissue for PCR Analysis
Purpose: To obtain high-quality genomic DNA for genotyping edited PDS loci. Materials: Liquid nitrogen, mortar and pestle, CTAB extraction buffer, chloroform:isoamyl alcohol (24:1), isopropanol, 70% ethanol, TE buffer, RNase A. Procedure:
- Harvest ~100 mg of leaf tissue from wild-type and putative edited plants (T0) and their progeny (T1, T2).
- Flash-freeze tissue in liquid nitrogen and grind to a fine powder.
- Transfer powder to a microtube with 500 µL pre-warmed (65°C) 2% CTAB buffer. Incubate at 65°C for 30 minutes with occasional mixing.
- Cool to room temperature. Add 500 µL chloroform:isoamyl alcohol, mix gently, and centrifuge at 12,000 g for 10 minutes.
- Transfer aqueous phase to a new tube. Add 0.7 volumes of isopropanol to precipitate DNA. Incubate at -20°C for 30 minutes.
- Pellet DNA by centrifugation (12,000 g, 10 min). Wash pellet with 70% ethanol.
- Air-dry pellet and resuspend in 50 µL TE buffer containing 5 µg/mL RNase A. Incubate at 37°C for 15 minutes.
- Quantify DNA using a spectrophotometer and store at -20°C.
Protocol: PCR Amplification and Sanger Sequencing ofPDSTarget Region
Purpose: To amplify the genomic region surrounding the CRISPR/Cas9 target site for sequencing analysis of edits. Materials: High-fidelity DNA polymerase, gene-specific primers flanking the target site, dNTPs, PCR purification kit. Procedure:
- Design primers ~300-500 bp upstream and downstream of the target site. Verify specificity via in silico PCR.
- Set up 25 µL PCR reaction: 50 ng genomic DNA, 1X polymerase buffer, 0.2 mM dNTPs, 0.5 µM each primer, 1 unit high-fidelity polymerase.
- Cycling conditions: Initial denaturation 98°C, 30 sec; 35 cycles of [98°C 10 sec, 60°C (or primer Tm) 15 sec, 72°C 30 sec/kb]; final extension 72°C, 2 min.
- Analyze 5 µL PCR product on a 1% agarose gel.
- Purify the remaining PCR product using a spin-column kit.
- Submit purified amplicon for Sanger sequencing using the forward or reverse PCR primer.
- Analyze sequencing chromatograms using alignment software (e.g., SnapGene, TIDE) to detect indels relative to the wild-type sequence.
Protocol: Segregation Analysis in T1 and T2 Progeny
Purpose: To determine the inheritance pattern of the edited PDS allele and identify transgene-free, homozygous plants. Materials: DNA from T1/T2 seedlings, PCR reagents for PDS amplification and Cas9 detection, agarose gel equipment. Procedure:
- Germination: Surface sterilize T1 seeds from self-pollinated T0 plants. Germinate on MS medium. Collect leaf tissue from ~20-30 individual seedlings.
- Genotyping: Extract gDNA from each seedling. Perform two parallel PCRs: a. Edit Detection: Amplify the PDS target region (Protocol 2.2). Sequence products or use a restriction enzyme assay (if edit disrupts a site) to determine genotype (Wild-type/WT, Heterozygous/Het, Homozygous Mutant/Hom). b. Cas9 Presence Detection: Amplify a region of the Cas9 transgene using specific primers.
- Segregation Scoring: Tally the number of T1 plants in each genotypic class (WT, Het, Hom) for the PDS edit and their Cas9 status (+/-).
- Chi-Square Test: Compare observed segregation ratios to expected Mendelian ratios (e.g., 1:2:1 for a heterozygous T0 edit, or 3:1 for a dominant phenotype) to assess statistical fit.
- Advancement: Select homozygous, Cas9-free T1 plants. Self-pollinate to generate T2 seeds. Repeat genotyping on a T2 population to confirm stable, transgene-free inheritance without further segregation.
Data Presentation
Table 1: Segregation Analysis of CRISPR/Cas9-InducedPDSEdits in T1 Progeny
| T0 Plant ID | T0 Genotype | # T1 Plants Screened | # WT | # Heterozygous | # Homozygous | Observed Ratio (WT:Het:Hom) | χ² (vs 1:2:1) | p-value | Cas9-free Homozygous Lines |
|---|---|---|---|---|---|---|---|---|---|
| PDS-01 | Heterozygous | 24 | 5 | 13 | 6 | 5:13:6 | 0.72 | >0.05 | 4 |
| PDS-07 | Heterozygous | 22 | 8 | 10 | 4 | 8:10:4 | 2.36 | >0.05 | 3 |
| PDS-12 | Biallelic | 20 | 0 | 7 | 13 | 0:7:13 | N/A* | N/A | 10 |
*Biallelic edits do not follow a simple 1:2:1 ratio. Expected ratio depends on specific T0 allele combination.
Table 2: Stability ofPDSEdits and Phenotype in Advanced Generations
| Plant Line (Origin) | Generation | Genotype (PDS) | Cas9 Transgene | Phenotype (Bleaching) | Edit Sequence (vs WT) |
|---|---|---|---|---|---|
| WT | - | WT/WT | - | Green | - |
| PDS-01-08 | T1 | Hom (Del4/Del4) | - | Bleached | -4 bp deletion |
| PDS-01-08 | T2 (n=15) | Hom (Del4/Del4) | - | Bleached (15/15) | -4 bp deletion |
| PDS-12-04 | T1 | Hom (Ins1/Del2) | + | Bleached | +1 bp / -2 bp |
| PDS-12-04 | T2 (n=15) | Segregating | Segregating | Segregating | NA |
Visualization Diagrams
Diagram Title: Heritability and Segregation Analysis Workflow for PDS Edits
Diagram Title: Mendelian Inheritance of a Heterozygous T0 Edit
The Scientist's Toolkit: Key Research Reagent Solutions
| Item/Category | Specific Example/Product | Function in PDS Editing/Heritability Analysis |
|---|---|---|
| CRISPR/Cas9 System | Agrobacterium strain GV3101 with binary vector (e.g., pCambia-Ubi:Cas9-PDSsgRNA) | Delivery of Cas9 and PDS-targeting sgRNA into plant cells. |
| Plant Selection Agent | Hygromycin B or Kanamycin (concentration varies by species) | Selection of transformed plant tissue carrying the CRISPR/Cas9 transgene. |
| High-Fidelity Polymerase | Q5 High-Fidelity DNA Polymerase (NEB) or Phusion Polymerase (Thermo) | Accurate PCR amplification of the PDS target region for sequencing. |
| Genotyping & Sequencing Kit | Sanger Sequencing Service with BigDye Terminator v3.1 (Thermo) | Determines the exact DNA sequence at the target locus to identify indels. |
| DNA Analysis Software | TIDE (Tracking of Indels by Decomposition) or ICE (Inference of CRISPR Edits) by Synthego | Deconvolutes Sanger sequencing chromatograms to quantify editing efficiency. |
| Gel Extraction/PCR Cleanup Kit | QIAquick Gel Extraction Kit or Monarch PCR & DNA Cleanup Kit (NEB) | Purifies DNA fragments for clean sequencing results or cloning. |
| Plant DNA Extraction Kit | DNeasy Plant Mini Kit (QIAGEN) or CTAB-based homemade reagents | Reliable isolation of PCR-ready genomic DNA from leaf tissue. |
| Agarose Gel Electrophoresis System | Standard horizontal gel system with SYBR Safe DNA stain | Visualizes PCR products for size verification and Cas9 transgene detection. |
Conclusion
CRISPR/Cas9-mediated editing of the phytoene desaturase (PDS) gene remains an indispensable, dual-purpose tool in plant biotechnology. It serves as a robust visual and molecular benchmark for optimizing genome editing pipelines while simultaneously unlocking research into carotenoid metabolism with direct implications for nutritional enhancement and abiotic stress tolerance. The methodologies and troubleshooting frameworks discussed enable researchers to achieve high-efficiency, specific knockouts. Looking forward, the validated PDS editing systems provide a foundational platform for more complex multiplexed editing and metabolic engineering. For biomedical and clinical research, this work is pivotal, as engineered plant lines with altered carotenoid pathways can become sustainable bioreactors for producing high-value apocarotenoids and isoprenoid-derived pharmaceuticals, bridging plant functional genomics with therapeutic discovery.